7LPS
 
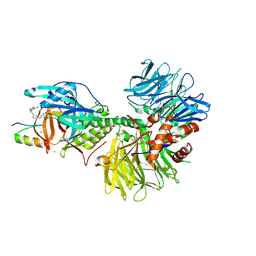 | | Crystal structure of DDB1-CRBN-ALV1 complex bound to Helios (IKZF2 ZF2) | | Descriptor: | 3-[3-[[1-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2,5-bis(oxidanylidene)pyrrol-3-yl]amino]phenyl]-~{N}-(3-chloranyl-4-methyl-phenyl)propanamide, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, Fischer, E.S. | | Deposit date: | 2021-02-12 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.78 Å) | | Cite: | Acute pharmacological degradation of Helios destabilizes regulatory T cells.
Nat.Chem.Biol., 17, 2021
|
|
6BN8
 
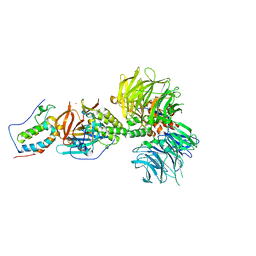 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET55 PROTAC. | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.990035 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6BNB
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET57 PROTAC | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Ishoey, M, He, Z, Zhang, T, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (6.343 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6BN9
 
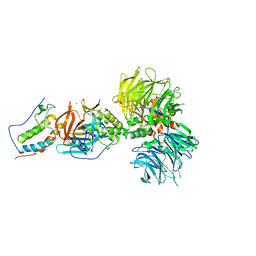 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET70 PROTAC | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.382 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6BN7
 
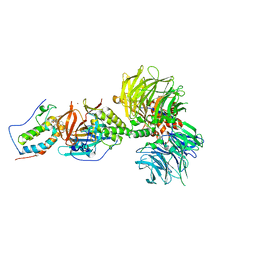 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET23 PROTAC. | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6BOY
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET6 PROTAC. | | Descriptor: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]-N-(8-{[({2-[(3S)-2,6-dioxopiperidin-3-yl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}oxy)acetyl]amino}octyl)acetamide, Bromodomain-containing protein 4, DNA damage-binding protein 1, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-21 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
7U20
 
 | | Crystal structure of human METTL1 and WDR4 complex | | Descriptor: | SULFATE ION, tRNA (guanine-N(7)-)-methyltransferase, tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4 | | Authors: | Li, J, Nowak, R.P, Fischer, E.S, Gregory, R. | | Deposit date: | 2022-02-22 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of regulated m 7 G tRNA modification by METTL1-WDR4.
Nature, 613, 2023
|
|
8UWB
 
 | | Crystal structure of PP2A PPP2R1A-PPP2CA-PPP2R5E phosphatase. | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit epsilon isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Wachter, F, Nowak, R.P, Fischer, E.S. | | Deposit date: | 2023-11-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural characterization of methylation-independent PP2A assembly guides alphafold2Multimer prediction of family-wide PP2A complexes.
J.Biol.Chem., 300, 2024
|
|
6XMX
 
 | | Cryo-EM structure of BCL6 bound to BI-3802 | | Descriptor: | 2-[6-[[5-chloranyl-2-[(3~{S},5~{R})-3,5-dimethylpiperidin-1-yl]pyrimidin-4-yl]amino]-1-methyl-2-oxidanylidene-quinolin-3-yl]oxy-~{N}-methyl-ethanamide, B-cell lymphoma 6 protein | | Authors: | Yoon, H, Burman, S.S.R, Hunkeler, M, Nowak, R.P, Fischer, E.S. | | Deposit date: | 2020-07-01 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Small-molecule-induced polymerization triggers degradation of BCL6.
Nature, 588, 2020
|
|
6Q0R
 
 | | Structure of DDB1-DDA1-DCAF15 complex bound to E7820 and RBM39 | | Descriptor: | 3-cyano-N-(3-cyano-4-methyl-1H-indol-7-yl)benzene-1-sulfonamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
6Q0W
 
 | | Structure of DDB1-DDA1-DCAF15 complex bound to Indisulam and RBM39 | | Descriptor: | DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1, ... | | Authors: | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
6Q0V
 
 | | Structure of DDB1-DDA1-DCAF15 complex bound to tasisulam and RBM39 | | Descriptor: | DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1, ... | | Authors: | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
5OXG
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with LDN-212854 | | Descriptor: | 1,2-ETHANEDIOL, 5-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Activin receptor type-1, ... | | Authors: | Williams, E.P, Sorrell, F.J, Kopec, J, Nowak, R.P, Kupinska, K, von Delft, F, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-06 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis for the potent and selective binding of LDN-212854 to the BMP receptor kinase ALK2.
Bone, 109, 2018
|
|
