1TUB
 
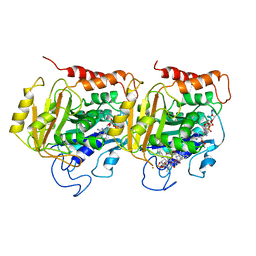 | | TUBULIN ALPHA-BETA DIMER, ELECTRON DIFFRACTION | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, TAXOTERE, ... | | Authors: | Nogales, E, Downing, K.H. | | Deposit date: | 1997-09-23 | | Release date: | 1998-10-07 | | Last modified: | 2024-06-05 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.7 Å) | | Cite: | Structure of the alpha beta tubulin dimer by electron crystallography.
Nature, 391, 1998
|
|
6CVN
 
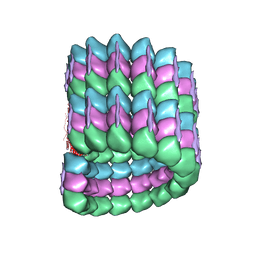 | |
6CVJ
 
 | |
6B5B
 
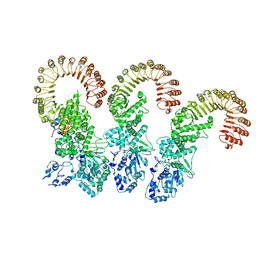 | | Cryo-EM structure of the NAIP5-NLRC4-flagellin inflammasome | | Descriptor: | Baculoviral IAP repeat-containing protein 1e, Flagellin, NLR family CARD domain-containing protein 4 | | Authors: | Tenthorey, J.L, Haloupek, N, Lopez-Blanco, J.R, Grob, P, Adamson, E, Hartenian, E, Lind, N.A, Bourgeois, N.M, Chacon, P, Nogales, E, Vance, R.E. | | Deposit date: | 2017-09-29 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | The structural basis of flagellin detection by NAIP5: A strategy to limit pathogen immune evasion.
Science, 358, 2017
|
|
1JFF
 
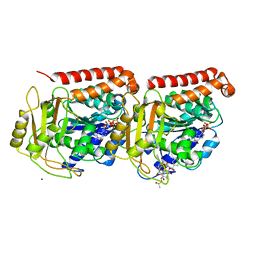 | | Refined structure of alpha-beta tubulin from zinc-induced sheets stabilized with taxol | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lowe, J, Li, H, Downing, K.H, Nogales, E. | | Deposit date: | 2001-06-20 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Refined structure of alpha beta-tubulin at 3.5 A resolution.
J.Mol.Biol., 313, 2001
|
|
8FIY
 
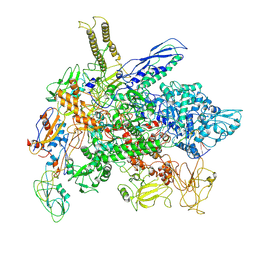 | | Cryo-EM structure of E. coli RNA polymerase Elongation complex in the Transcription-Translation Complex (RNAP in an anti-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Florez Ariza, A, Wee, L, Tong, A, Canari, C, Grob, P, Nogales, E, Bustamante, C. | | Deposit date: | 2022-12-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | A trailing ribosome speeds up RNA polymerase at the expense of transcript fidelity via force and allostery.
Cell, 186, 2023
|
|
8FIX
 
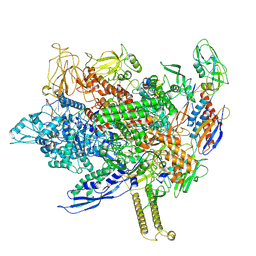 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex harboring a terminal mismatch | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Florez Ariza, A, Wee, L, Tong, A, Canari, C, Grob, P, Nogales, E, Bustamante, C. | | Deposit date: | 2022-12-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A trailing ribosome speeds up RNA polymerase at the expense of transcript fidelity via force and allostery.
Cell, 186, 2023
|
|
8FIZ
 
 | | Cryo-EM structure of E. coli 70S Ribosome containing mRNA and tRNA (in the transcription-translation complex) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Florez Ariza, A, Wee, L, Tong, A, Canari, C, Grob, P, Nogales, E, Bustamante, C. | | Deposit date: | 2022-12-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A trailing ribosome speeds up RNA polymerase at the expense of transcript fidelity via force and allostery.
Cell, 186, 2023
|
|
8T9G
 
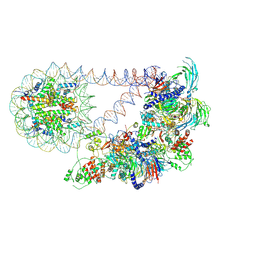 | | Automethylated PRC2 dimer bound to nucleosome | | Descriptor: | DNA (226-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Sauer, P.V, Pavlenko, E, Nogales, E, Poepsel, S. | | Deposit date: | 2023-06-23 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Activation of automethylated PRC2 by dimerization on chromatin.
Mol.Cell, 2024
|
|
8TB9
 
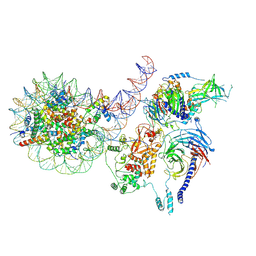 | | PRC2-J119-450 monomer bound to H1-nucleosome | | Descriptor: | DNA (226-MER), Histone H1.0, Histone H2A type 1, ... | | Authors: | Sauer, P.V, Cookis, T, Pavlenko, E, Nogales, E, Poepsel, S. | | Deposit date: | 2023-06-28 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Activation of automethylated PRC2 by dimerization on chromatin.
Mol.Cell, 2024
|
|
8TAS
 
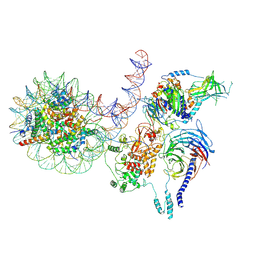 | | PRC2 monomer bound to nucleosome | | Descriptor: | DNA (226-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Sauer, P.V, Pavlenko, E, Nogales, E, Poepsel, S. | | Deposit date: | 2023-06-27 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Activation of automethylated PRC2 by dimerization on chromatin.
Mol.Cell, 2024
|
|
7B5Q
 
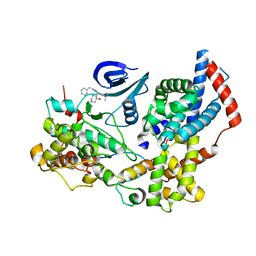 | | Cryo-EM structure of the human CAK bound to ICEC0942 (PHENIX-OPLS3e) | | Descriptor: | (3R,4R)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Greber, B.J, Remis, J, Ali, S, Nogales, E. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | 2.5 angstrom -resolution structure of human CDK-activating kinase bound to the clinical inhibitor ICEC0942.
Biophys.J., 120, 2021
|
|
7B5O
 
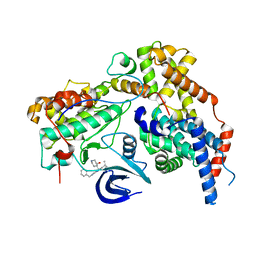 | | Cryo-EM structure of the human CAK bound to ICEC0942 at 2.5 Angstroms resolution | | Descriptor: | (3R,4R)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Greber, B.J, Remis, J, Ali, S, Nogales, E. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | 2.5 angstrom -resolution structure of human CDK-activating kinase bound to the clinical inhibitor ICEC0942.
Biophys.J., 120, 2021
|
|
5KMG
 
 | | Near-atomic cryo-EM structure of PRC1 bound to the microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kellogg, E.H, Howes, S, Ti, S.-C, Ramirez-Aportela, E, Kapoor, T.M, Chacon, P, Nogales, E. | | Deposit date: | 2016-06-27 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Near-atomic cryo-EM structure of PRC1 bound to the microtubule.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4CMQ
 
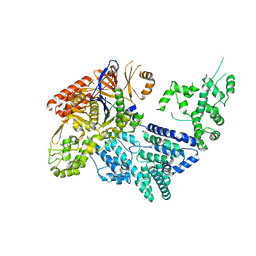 | | Crystal structure of Mn-bound S.pyogenes Cas9 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MANGANESE (II) ION, SULFATE ION | | Authors: | Jinek, M, Jiang, F, Taylor, D.W, Sternberg, S.H, Kaya, E, Ma, E, Anders, C, Hauer, M, Zhou, K, Lin, S, Kaplan, M, Iavarone, A.T, Charpentier, E, Nogales, E, Doudna, J.A. | | Deposit date: | 2014-01-17 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structures of Cas9 Endonucleases Reveal RNA- Mediated Conformational Activation
Science, 343, 2014
|
|
4CMP
 
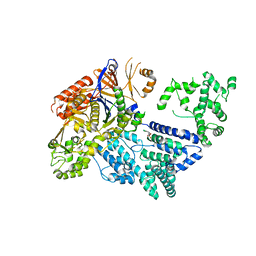 | | Crystal structure of S. pyogenes Cas9 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, SULFATE ION | | Authors: | Jinek, M, Jiang, F, Taylor, D.W, Sternberg, S.H, Kaya, E, Ma, E, Anders, C, Hauer, M, Zhou, K, Lin, S, Kaplan, M, Iavarone, A.T, Charpentier, E, Nogales, E, Doudna, J.A. | | Deposit date: | 2014-01-16 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation.
Science, 343, 2014
|
|
6X8M
 
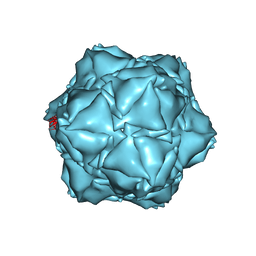 | | CryoEM structure of the holo-SrpI encapsulin complex from Synechococcus elongatus PCC 7942 | | Descriptor: | Protein SrpI | | Authors: | LaFrance, B.J, Nichols, R.J, Phillips, N.R, Oltrogge, L.M, Valentin-Alvarado, L.E, Bischoff, A.J, Savage, D.F, Nogales, E. | | Deposit date: | 2020-06-01 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Discovery and characterization of a novel family of prokaryotic nanocompartments involved in sulfur metabolism.
Elife, 10, 2021
|
|
6X8T
 
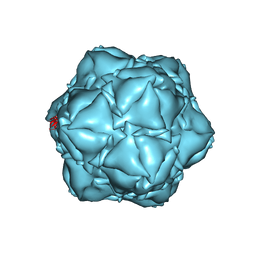 | | CryoEM structure of the apo-SrpI encapasulin complex from Synechococcus elongatus PCC 7942 | | Descriptor: | Protein SrpI | | Authors: | LaFrance, B.J, Nichols, R.J, Phillips, N.R, Oltrogge, L.M, Valentin-Alvarado, L.E, Bischoff, A.J, Savage, D.F, Nogales, E. | | Deposit date: | 2020-06-01 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Discovery and characterization of a novel family of prokaryotic nanocompartments involved in sulfur metabolism.
Elife, 10, 2021
|
|
5OF4
 
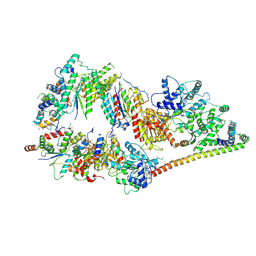 | | The cryo-EM structure of human TFIIH | | Descriptor: | General transcription factor IIH subunit 2, General transcription factor IIH subunit 3, General transcription factor IIH subunit 4,p52,General transcription factor IIH subunit 4, ... | | Authors: | Greber, B.J, Nguyen, T.H.D, Fang, J, Afonine, P.V, Adams, P.D, Nogales, E. | | Deposit date: | 2017-07-10 | | Release date: | 2017-09-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The cryo-electron microscopy structure of human transcription factor IIH.
Nature, 549, 2017
|
|
8ESC
 
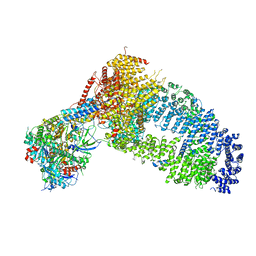 | | Structure of the Yeast NuA4 Histone Acetyltransferase Complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Patel, A.B, Zukin, S.A, Nogales, E. | | Deposit date: | 2022-10-13 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and flexibility of the yeast NuA4 histone acetyltransferase complex.
Elife, 11, 2022
|
|
6P5A
 
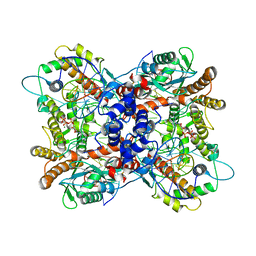 | | Drosophila P element transposase strand transfer complex | | Descriptor: | DNA (44-MER), DNA (5'-D(P*AP*GP*GP*TP*GP*GP*TP*CP*CP*CP*GP*TP*CP*GP*G)-3'), DNA (5'-D(P*CP*GP*AP*AP*CP*TP*AP*TP*A)-3'), ... | | Authors: | Kellogg, E.H, Nogales, E, Ghanim, G, Rio, D.C. | | Deposit date: | 2019-05-30 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a P element transposase-DNA complex reveals unusual DNA structures and GTP-DNA contacts.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6PE2
 
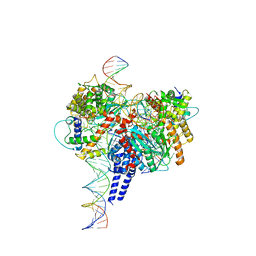 | | Drosophila P element transposase strand transfer complex | | Descriptor: | DNA (27-MER), DNA (5'-D(P*CP*GP*AP*AP*CP*TP*AP*TP*A)-3'), DNA (56-MER), ... | | Authors: | Kellogg, E.H, Nogales, E, Ghanim, G, Rio, D.C. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of a P element transposase-DNA complex reveals unusual DNA structures and GTP-DNA contacts.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5IYA
 
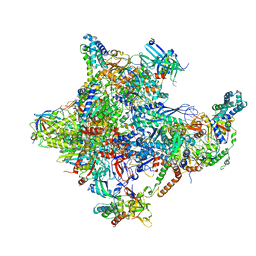 | | Human core-PIC in the closed state | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | Deposit date: | 2016-03-24 | | Release date: | 2016-05-18 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
7M5O
 
 | | Cryo-EM structure of CasPhi-2 (Cas12j) bound to crRNA | | Descriptor: | CasPhi, ZINC ION, crRNA | | Authors: | Pausch, P, Soczek, K, Nogales, E, Doudna, J. | | Deposit date: | 2021-03-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | DNA interference states of the hypercompact CRISPR-Cas Phi effector.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5IY8
 
 | | Human holo-PIC in the initial transcribing state | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | Deposit date: | 2016-03-24 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
