2OXQ
 
 | | Structure of the UbcH5 :CHIP U-box complex | | Descriptor: | CHLORIDE ION, STIP1 homology and U-Box containing protein 1, Ubiquitin-conjugating enzyme E2D 1 | | Authors: | Xu, Z, Nix, J.C, Devlin, K.I, Misra, S. | | Deposit date: | 2007-02-20 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes.
Bmc Struct.Biol., 8, 2008
|
|
6OP7
 
 | | Structure of oxidized VIM-20 | | Descriptor: | ACETATE ION, Metallo-beta-lactamase VIM-20, ZINC ION | | Authors: | Page, R.C, Shurina, B.A, Montgomery, J.S, Orischak, M.G, Nix, J.C. | | Deposit date: | 2019-04-24 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | A Single Salt Bridge in VIM-20 Increases Protein Stability and Antibiotic Resistance under Low-Zinc Conditions.
Mbio, 10, 2019
|
|
2F42
 
 | |
1DDY
 
 | |
4GKY
 
 | | Crystal structure of a carbohydrate-binding domain | | Descriptor: | CALCIUM ION, GLYCEROL, Protein ERGIC-53, ... | | Authors: | Page, R.C, Zheng, C, Nix, J.C, Misra, S, Zhang, B. | | Deposit date: | 2012-08-13 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4201 Å) | | Cite: | Structural Characterization of Carbohydrate Binding by LMAN1 Protein Provides New Insight into the Endoplasmic Reticulum Export of Factors V (FV) and VIII (FVIII).
J.Biol.Chem., 288, 2013
|
|
4GKX
 
 | | Crystal structure of a carbohydrate-binding domain | | Descriptor: | CALCIUM ION, GLYCEROL, Protein ERGIC-53, ... | | Authors: | Page, R.C, Zheng, C, Nix, J.C, Misra, S, Zhang, B. | | Deposit date: | 2012-08-13 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Characterization of Carbohydrate Binding by LMAN1 Protein Provides New Insight into the Endoplasmic Reticulum Export of Factors V (FV) and VIII (FVIII).
J.Biol.Chem., 288, 2013
|
|
9YD7
 
 | | Complex of Dihydroorotase from M. jannaschii with Carbamoyl Aspartate | | Descriptor: | Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ZINC ION | | Authors: | Vitali, J, Nix, J.C, Newman, H.E, Colaneri, M.J. | | Deposit date: | 2025-09-22 | | Release date: | 2025-12-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of Methanococcus jannaschii dihydroorotase with substrate bound.
Acta Crystallogr.,Sect.F, 2026
|
|
9YEA
 
 | |
9YE9
 
 | | Structure of UbcH5b in complex with the U-box domain of the E3 ubiquitin ligase CHIP | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, E3 ubiquitin-protein ligase CHIP, ... | | Authors: | Manage, M.M, Nix, J.C, Page, R.C. | | Deposit date: | 2025-09-23 | | Release date: | 2025-11-05 | | Last modified: | 2025-11-12 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Functional and structural analyses of UbcH5 mutants with enhanced binding to the E3 ubiquitin ligase CHIP.
Biochem.Biophys.Res.Commun., 789, 2025
|
|
8D3F
 
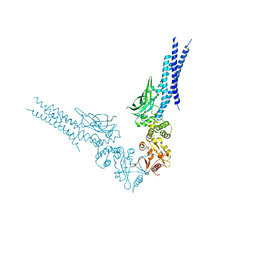 | | Crystal structure of human STAT1 in complex with the repeat region from Toxoplasma protein TgIST | | Descriptor: | Signal transducer and activator of transcription 1-alpha/beta,Inhibitor of STAT1-dependent transcription TgIST | | Authors: | Huang, Z, Liu, H, Nix, J.C, Amarasinghe, G.K, Sibley, L.D. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | The intrinsically disordered protein TgIST from Toxoplasma gondii inhibits STAT1 signaling by blocking cofactor recruitment.
Nat Commun, 13, 2022
|
|
7R6X
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2E12 Fab, S309 Fab, and S304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Monoclonal antibody S2E12 Fab heavy chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
6PMT
 
 | |
5TPY
 
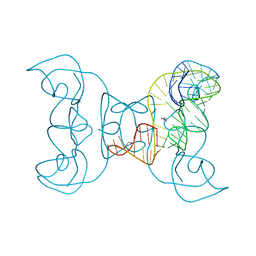 | | Crystal structure of an exonuclease resistant RNA from Zika virus | | Descriptor: | HEXANE-1,6-DIOL, MAGNESIUM ION, RNA (71-MER) | | Authors: | Akiyama, B.M, Laurence, H.M, Massey, A.R, Costantino, D.A, Xie, X, Yang, Y, Shi, P.-Y, Nix, J.C, Beckham, J.D, Kieft, J.S. | | Deposit date: | 2016-10-21 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease.
Science, 354, 2016
|
|
8TT9
 
 | | X-ray structure of Macrophage Migration Inhibitory Factor (MIF) Covalently Bound to 4-hydroxyphenylpyruvate (HPP) | | Descriptor: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor | | Authors: | Schroder, G.C, Meilleur, F, Nix, J.C, Crichlow, G.V, Lolis, E.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-08-28 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray structure of Macrophage Migration Inhibitory Factor (MIF) Covalently Bound to 4-hydroxyphenylpyruvate (HPP)
To Be Published
|
|
8UHL
 
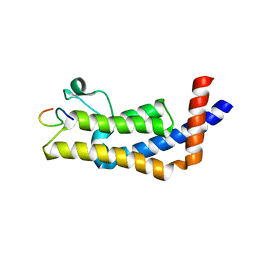 | | ATAD2B bromodomain in complex with histone H4 acetylated at lysine 12 | | Descriptor: | ATPase family AAA domain-containing protein 2B, Histone H4 | | Authors: | Phillips, M, Montgomery, C, Nix, J.C, Glass, K.C. | | Deposit date: | 2023-10-09 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Impact of Combinatorial Histone Modifications on Acetyllysine Recognition by the ATAD2 and ATAD2B Bromodomains.
J.Med.Chem., 67, 2024
|
|
8UK5
 
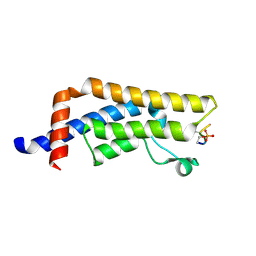 | | Crystal structure of the bromodomain of human ATAD2B in complex with histone H4S1(ph)K5ac | | Descriptor: | ATPase family AAA domain-containing protein 2B, Histone H4S1(ph)K5ac | | Authors: | Montgomery, C, Phillips, M, Nix, J.C, Glass, K.C. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Impact of Combinatorial Histone Modifications on Acetyllysine Recognition by the ATAD2 and ATAD2B Bromodomains.
J.Med.Chem., 67, 2024
|
|
2QVV
 
 | | Porcine Liver Fructose-1,6-bisphosphatase cocrystallized with Fru-2,6-P2 and Zn2+, I(T)-state | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase 1, PHOSPHATE ION, ... | | Authors: | Hines, J.K, Chen, X, Nix, J.C, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of mammalian and bacterial fructose-1,6-bisphosphatase reveal the basis for synergism in AMP/fructose 2,6-bisphosphate inhibition
J.Biol.Chem., 282, 2007
|
|
2QVU
 
 | | Porcine Liver Fructose-1,6-bisphosphatase cocrystallized with Fru-2,6-P2 and Mg2+, I(T)-state | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase 1, MAGNESIUM ION, ... | | Authors: | Hines, J.K, Chen, X, Nix, J.C, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of mammalian and bacterial fructose-1,6-bisphosphatase reveal the basis for synergism in AMP/fructose 2,6-bisphosphate inhibition
J.Biol.Chem., 282, 2007
|
|
7UOF
 
 | | Dihydroorotase from M. jannaschii | | Descriptor: | Dihydroorotase, ZINC ION | | Authors: | Vitali, J, Nix, J.C, Newman, H.E, Colaneri, M.J. | | Deposit date: | 2022-04-12 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Methanococcus jannaschii dihydroorotase.
Proteins, 91, 2023
|
|
7R6W
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2X35 Fab and S309 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Snell, G, Czudnochowski, N, Hernandez, P, Nix, J.C, Croll, T.I, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
7TN0
 
 | | SARS-CoV-2 Omicron RBD in complex with human ACE2 and S304 Fab and S309 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | McCallum, M, Czudnochowski, N, Nix, J.C, Croll, T.I, SSGCID, Dillen, J.R, Snell, G, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement.
Science, 375, 2022
|
|
8SDQ
 
 | |
8SDO
 
 | |
8SDX
 
 | | ATAD2B bromodomain in complex with histone H4 acetylated at lysine 5 with Serine 1 mutation to Cysteine | | Descriptor: | ATPase family AAA domain-containing protein 2B, SULFATE ION, histone H4S1CK5ac | | Authors: | Phillips, M, Montgomery, C, Nix, J.C, Glass, K.C. | | Deposit date: | 2023-04-07 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Impact of Combinatorial Histone Modifications on Acetyllysine Recognition by the ATAD2 and ATAD2B Bromodomains.
J.Med.Chem., 67, 2024
|
|
3CQX
 
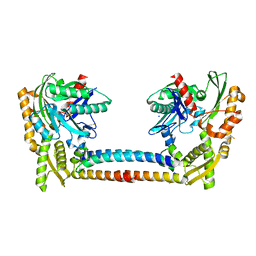 | | Chaperone Complex | | Descriptor: | BAG family molecular chaperone regulator 2, Heat shock cognate 71 kDa protein, SODIUM ION, ... | | Authors: | Xu, Z, Nix, J.C, Misra, S. | | Deposit date: | 2008-04-03 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of nucleotide exchange and client binding by the Hsp70 cochaperone Bag2
Nat.Struct.Mol.Biol., 15, 2008
|
|
