3NKM
 
 | | Crystal structure of mouse autotaxin | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NKN
 
 | | Crystal structure of mouse autotaxin in complex with 14:0-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NKQ
 
 | | Crystal structure of mouse autotaxin in complex with 18:3-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E,12Z,15Z)-octadeca-9,12,15-trienoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NKO
 
 | | Crystal structure of mouse autotaxin in complex with 16:0-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NKP
 
 | | Crystal structure of mouse autotaxin in complex with 18:1-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NKR
 
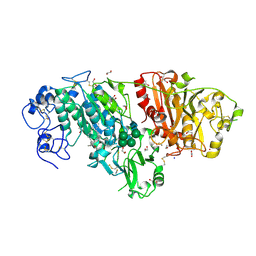 | | Crystal structure of mouse autotaxin in complex with 22:6-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (4Z,7E,10E,13Z,16Z,19Z)-docosa-4,7,10,13,16,19-hexaenoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
5AXW
 
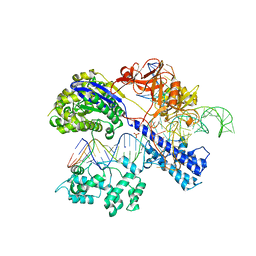 | | Crystal structure of Staphylococcus aureus Cas9 in complex with sgRNA and target DNA (TTGGGT PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, DNA (28-MER), ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2015-08-01 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Staphylococcus aureus Cas9.
Cell, 162, 2015
|
|
4YBG
 
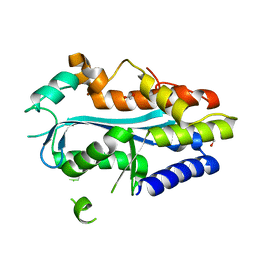 | | Crystal structure of the MAEL domain of Drosophila melanogaster Maelstrom | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Protein maelstrom, ... | | Authors: | Matsumoto, N, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Crystal Structure and Activity of the Endoribonuclease Domain of the piRNA Pathway Factor Maelstrom
Cell Rep, 11, 2015
|
|
8GS2
 
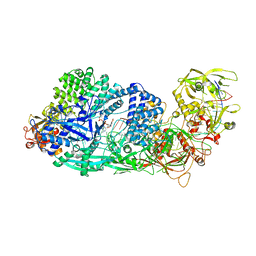 | | Structure of the Cas7-11-Csx29-guide RNA-target RNA (non-matching PFS) complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHAT domain-containing protein, CRISPR-associated RAMP family protein, ... | | Authors: | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-09-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
5XUZ
 
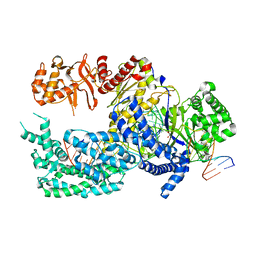 | | Crystal structure of Lachnospiraceae bacterium ND2006 Cpf1 in complex with crRNA and target DNA (CCCA PAM) | | Descriptor: | 1,2-ETHANEDIOL, DNA (29-MER), DNA (5'-D(*CP*GP*TP*CP*CP*CP*CP*CP*A)-3'), ... | | Authors: | Yamano, T, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-26 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Mol. Cell, 67, 2017
|
|
7C7L
 
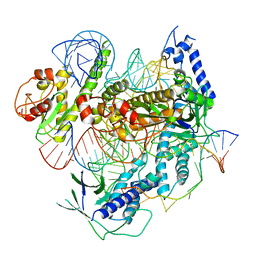 | | Cryo-EM structure of the Cas12f1-sgRNA-target DNA complex | | Descriptor: | CRISPR-associated protein Cas14a.1, DNA (40-mer), ZINC ION, ... | | Authors: | Takeda, N.S, Nakagawa, R, Okazaki, S, Hirano, H, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2020-05-26 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the miniature type V-F CRISPR-Cas effector enzyme.
Mol.Cell, 81, 2021
|
|
5JYJ
 
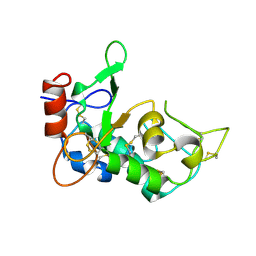 | | Crystal structure of mouse JUNO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sperm-egg fusion protein Juno | | Authors: | Kato, K, Nishimasu, H, Morita, J, Ishitani, R, Nureki, O. | | Deposit date: | 2016-05-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the egg IZUMO1 receptor JUNO
To Be Published
|
|
5XUT
 
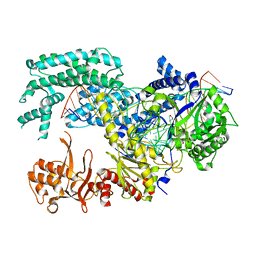 | | Crystal structure of Lachnospiraceae bacterium ND2006 Cpf1 in complex with crRNA and target DNA (TCTA PAM) | | Descriptor: | 1,2-ETHANEDIOL, DNA (29-MER), DNA (5'-D(*CP*GP*TP*CP*CP*TP*CP*TP*A)-3'), ... | | Authors: | Yamano, T, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-26 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Mol. Cell, 67, 2017
|
|
6IS0
 
 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH and m7G-capped RNA | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
4L3O
 
 | | Crystal Structure of SIRT2 in complex with the macrocyclic peptide S2iL5 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Yamagata, K, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2013-06-06 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.518 Å) | | Cite: | Structural Basis for Potent Inhibition of SIRT2 Deacetylase by a Macrocyclic Peptide Inducing Dynamic Structural Change
Structure, 22, 2013
|
|
3R1M
 
 | | Structure of bifunctional fructose 1,6-bisphosphate aldolase/phosphatase (aldolase form) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fushinobu, S, Nishimasu, H, Hattori, D, Song, H.-J, Wakagi, T. | | Deposit date: | 2011-03-10 | | Release date: | 2011-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the bifunctionality of fructose-1,6-bisphosphate aldolase/phosphatase.
Nature, 478, 2011
|
|
7Y9Y
 
 | | Structure of the Cas7-11-Csx29-guide RNA-target RNA (no PFS) complex | | Descriptor: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, RNA (27-MER), ... | | Authors: | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-06-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
6IRZ
 
 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH and m7G-capped RNA | | Descriptor: | 1,2-ETHANEDIOL, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, PDX1 C-terminal-inhibiting factor 1, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6IRV
 
 | |
7Y9X
 
 | | Structure of the Cas7-11-Csx29-guide RNA complex | | Descriptor: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, ZINC ION, ... | | Authors: | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-06-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
5X2H
 
 | | Crystal structure of Campylobacter jejuni Cas9 in complex with sgRNA and target DNA (AGAAACA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, Non-target DNA strand, ... | | Authors: | Yamada, M, Watanabe, Y, Hirano, H, Nakane, T, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems
Mol. Cell, 65, 2017
|
|
5X2G
 
 | | Crystal structure of Campylobacter jejuni Cas9 in complex with sgRNA and target DNA (AGAAACC PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, Non-target DNA strand, ... | | Authors: | Yamada, M, Watanabe, Y, Hirano, H, Nakane, T, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems
Mol. Cell, 65, 2017
|
|
5EGH
 
 | | Structure of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase in complex with phosphocholine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Morita, J, Kano, K, Kato, K, Takita, H, Ishitani, R, Nishimasu, H, Nureki, O, Aoki, J. | | Deposit date: | 2015-10-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structure and biological function of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase
Sci Rep, 6, 2016
|
|
5EGE
 
 | | Structure of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Morita, J, Kano, K, Kato, K, Takita, H, Ishitani, R, Nishimasu, H, Nureki, O, Aoki, J. | | Deposit date: | 2015-10-27 | | Release date: | 2016-03-09 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biological function of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase
Sci Rep, 6, 2016
|
|
5GIJ
 
 | | Crystal structure of TDR-TDIF complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeat receptor-like protein kinase TDR, ... | | Authors: | Morita, J, Kato, K, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2016-06-23 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the plant receptor-like kinase TDR in complex with the TDIF peptide
Nat Commun, 7, 2016
|
|
