5XVK
 
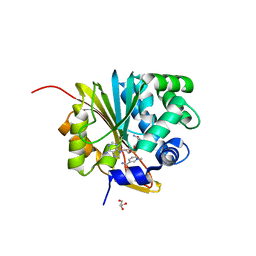 | | Crystal structure of mouse Nicotinamide N-methyltransferase (NNMT) bound with end product, 1-methyl Nicotinamide (MNA) | | Descriptor: | 3-carbamoyl-1-methylpyridin-1-ium, GLYCEROL, Nicotinamide N-methyltransferase, ... | | Authors: | Swaminathan, S, Birudukota, S, Thakur, M.K, Parveen, R, Kandan, S, Kannt, A, Gosu, R. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of monkey and mouse nicotinamide N-methyltransferase (NNMT) bound with end product, 1-methyl nicotinamide
Biochem. Biophys. Res. Commun., 491, 2017
|
|
2G5U
 
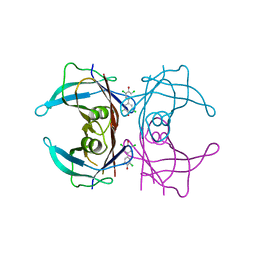 | | Human Transthyretin (TTR) Complexed with Hydroxylated polychlorinated Biphenyl-4,4'-dihydroxy-3,3',5,5'-tetrachlorobiphenyl | | Descriptor: | 3,5,3',5'-TETRACHLORO-BIPHENYL-4,4'-DIOL, Transthyretin | | Authors: | Palaninathan, S.K, Smith, C, Safe, S.H, Kelly, J.W, Sacchettini, J.C. | | Deposit date: | 2006-02-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hydroxylated polychlorinated biphenyls selectively bind transthyretin in blood and inhibit amyloidogenesis: rationalizing rodent PCB toxicity
Chem.Biol., 11, 2004
|
|
2G9K
 
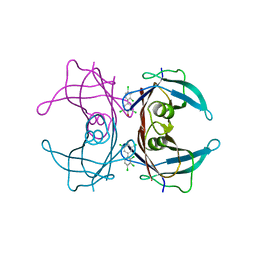 | | Human Transthyretin (TTR) Complexed with Hydroxylated polychlorinated Biphenyl-4-hydroxy-2',3,3',4',5-Pentachlorobiphenyl | | Descriptor: | 2',3,3',4',5-PENTACHLOROBIPHENYL-4-OL, Transthyretin | | Authors: | Palaninathan, S.K, Smith, C, Safe, S.H, Kelly, J.W, Sacchettini, J.C. | | Deposit date: | 2006-03-06 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Hydroxylated polychlorinated biphenyls selectively bind transthyretin in blood and inhibit amyloidogenesis: rationalizing rodent PCB toxicity
Chem.Biol., 11, 2004
|
|
5YJF
 
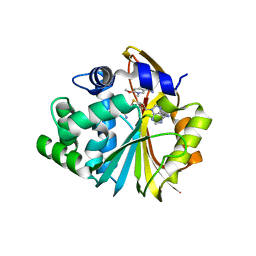 | | Co-crystal structure of Human Nicotinamide N-methyltransferase (NNMT) with small molecule analog of Nicotinamide | | Descriptor: | 6-methoxy-1-methyl-2H-pyridine-3-carboxamide, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Swaminathan, S, Birudukota, S, Thakur, M.K, Parveen, R, Kandan, S, Hallur, M.S, Rajagopal, S, Ruf, S, Dhakshinamoorthy, S, Kannt, A, Gosu, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | A small molecule inhibitor of Nicotinamide N-methyltransferase for the treatment of metabolic disorders.
Sci Rep, 8, 2018
|
|
1HNB
 
 | | CRYSTAL STRUCTURE OF HUMAN CLASS MU GLUTATHIONE TRANSFERASE GSTM2-2: EFFECTS OF LATTICE PACKING ON CONFORMATIONAL HETEROGENEITY | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Raghunathan, S, Chandross, R.J, Kretsinger, R.H, Allison, T.J, Penington, C.J, Rule, G.S. | | Deposit date: | 1993-10-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of human class mu glutathione transferase GSTM2-2. Effects of lattice packing on conformational heterogeneity.
J.Mol.Biol., 238, 1994
|
|
1HNC
 
 | | CRYSTAL STRUCTURE OF HUMAN CLASS MU GLUTATHIONE TRANSFERASE GSTM2-2: EFFECTS OF LATTICE PACKING ON CONFORMATIONAL HETEROGENEITY | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Raghunathan, S, Chandross, R.J, Kretsinger, R.H, Allison, T.J, Penington, C.J, Rule, G.S. | | Deposit date: | 1993-10-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human class mu glutathione transferase GSTM2-2. Effects of lattice packing on conformational heterogeneity.
J.Mol.Biol., 238, 1994
|
|
7EU5
 
 | | Co-crystal structure of Human Nicotinamide N-methyltransferase (NNMT) with tricyclic small molecule inhibitor JBSNF-000107 | | Descriptor: | 6-fluoranyl-10-methyl-1,10-diazatricyclo[6.3.1.0^{4,12}]dodeca-4,6,8(12)-trien-11-imine, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Swaminathan, S, Gosu, R, Birudukota, S, Kandan, S, Vaithilingam, K. | | Deposit date: | 2021-05-16 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.731 Å) | | Cite: | Novel tricyclic small molecule inhibitors of Nicotinamide N-methyltransferase for the treatment of metabolic disorders.
Sci Rep, 12, 2022
|
|
7ET7
 
 | | Co-crystal structure of Human Nicotinamide N-methyltransferase (NNMT) with tricyclic small molecule inhibitor JBSNF-000028 | | Descriptor: | 10-methyl-1,10-diazatricyclo[6.3.1.0^{4,12}]dodeca-4,6,8(12)-trien-11-imine, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Swaminathan, S, Gosu, R, Birudukota, S, Kandan, S, Vaithilingam, K. | | Deposit date: | 2021-05-12 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Novel tricyclic small molecule inhibitors of Nicotinamide N-methyltransferase for the treatment of metabolic disorders.
Sci Rep, 12, 2022
|
|
2GAB
 
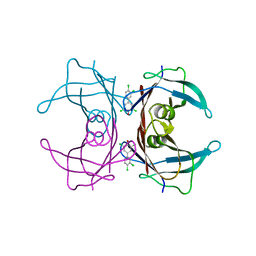 | | Human Transthyretin (TTR) Complexed with Hydroxylated polychlorinated Biphenyl-4-hydroxy-3,3',5,4'-tetrachlorobiphenyl | | Descriptor: | 3,3',4',5-TETRACHLOROBIPHENYL-4-OL, Transthyretin | | Authors: | Palaninathan, S.K, Smith, C, Safe, S.H, Kelly, J.W, Sacchettini, J.C. | | Deposit date: | 2006-03-08 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Hydroxylated polychlorinated biphenyls selectively bind transthyretin in blood and inhibit amyloidogenesis: rationalizing rodent PCB toxicity
Chem.Biol., 11, 2004
|
|
2ETF
 
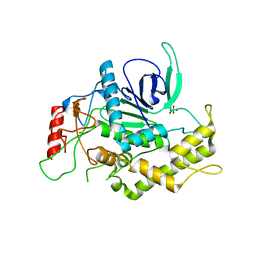 | |
2O8O
 
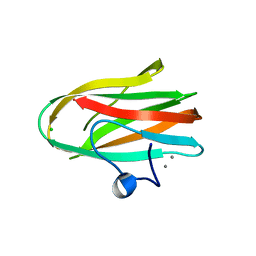 | |
6RVU
 
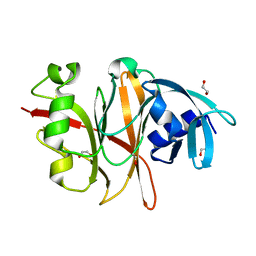 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) | | Descriptor: | 1,2-ETHANEDIOL, Lethal Factor 1 (BLF1) | | Authors: | Mobbs, G.W, Aziz, A.A, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | Deposit date: | 2019-06-01 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
7PQ0
 
 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) C94S inactive mutant in complex with human eIF4A - Crystal form B | | Descriptor: | Burkholderia Lethal Factor 1 (BLF1), Eukaryotic initiation factor 4A-I | | Authors: | Mobbs, G.W, Aziz, A.A, Dix, S.R, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | Deposit date: | 2021-09-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
7PPZ
 
 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) C94S inactive mutant in complex with human eIF4A - Crystal form A | | Descriptor: | Burkholderia Lethal Factor 1 (BLF1), Eukaryotic initiation factor 4A-I | | Authors: | Mobbs, G.W, Aziz, A.A, Dix, S.R, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | Deposit date: | 2021-09-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
7PZT
 
 | | Structure of the bacterial toxin, TecA, an asparagine deamidase from Alcaligenes faecalis. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Urea amidohydrolase | | Authors: | Dix, S.R, Aziz, A.A, Baker, P.J, Evans, C.A, Dickman, M.J, Farthing, R.J, King, Z.L.S, Nathan, S, Partridge, L.J, Raih, F.M, Sedelnikova, S.E, Thomas, M.S, Rice, D.W. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The structure of A. faecalis TecA provides insights into its role as an asparagine deamidase toxin which targets RhoA
To Be Published
|
|
7VXT
 
 | | Crystal structure of a selenomethionine-labeled BPSL1038 from Burkholderia pseudomallei | | Descriptor: | BETA-MERCAPTOETHANOL, BPSL1038, SODIUM ION | | Authors: | Shaibullah, S, Mohd-Sharif, M, Ho, K.L, Firdaus-Raih, M, Nathan, S, Mohamed, R, Teh, A.K, Waterman, J, Ng, C.L. | | Deposit date: | 2021-11-13 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and functional analyses of Burkholderia pseudomallei BPSL1038 reveal a Cas-2/VapD nuclease sub-family.
Commun Biol, 6, 2023
|
|
7VXR
 
 | | Crystal structure of BPSL1038 from Burkholderia pseudomallei | | Descriptor: | BPSL1038, SODIUM ION | | Authors: | Shaibullah, S, Mohd-Sharif, M, Ho, K.L, Firdaus-Raih, M, Nathan, S, Mohamed, R, Teh, A.K, Waterman, J, Ng, C.L. | | Deposit date: | 2021-11-13 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and functional analyses of Burkholderia pseudomallei BPSL1038 reveal a Cas-2/VapD nuclease sub-family.
Commun Biol, 6, 2023
|
|
3TUA
 
 | | Crystal Structure of the Burkholderia Lethal Factor 1 (BLF1) C94S mutant | | Descriptor: | Burkholderia Lethal Factor 1 (BLF1) | | Authors: | Cruz, A, Hautbergue, G.M, Artymiuk, P.J, Baker, P.J, Chang, C.T, Mahadi, N.M, Mobbs, G.W, Mohamed, R, Nathan, S, Partridge, L.J, Raih, M.F, Ruzheinikov, S.N, Sedelnikova, S.E, Wilson, S.A, Rice, D.W. | | Deposit date: | 2011-09-16 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | A Burkholderia pseudomallei toxin inhibits helicase activity of translation factor eIF4A.
Science, 334, 2011
|
|
3TU8
 
 | | Crystal Structure of the Burkholderia Lethal Factor 1 (BLF1) | | Descriptor: | BROMIDE ION, Burkholderia Lethal Factor 1 (BLF1) | | Authors: | Cruz, A, Hautbergue, G.M, Artymiuk, P.J, Baker, P.J, Chang, C.T, Mahadi, N.M, Mobbs, G.W, Mohamed, R, Nathan, S, Partridge, L.J, Raih, M.F, Ruzheinikov, S.N, Sedelnikova, S.E, Wilson, S.A, Rice, D.W. | | Deposit date: | 2011-09-16 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | A Burkholderia pseudomallei toxin inhibits helicase activity of translation factor eIF4A.
Science, 334, 2011
|
|
3DCN
 
 | | Glomerella cingulata apo cutinase | | Descriptor: | Cutinase | | Authors: | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | Deposit date: | 2008-06-04 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
3DEA
 
 | | Glomerella cingulata PETFP-cutinase complex | | Descriptor: | 1,1,1-trifluoro-3-[(2-phenylethyl)sulfanyl]propan-2-one, Cutinase | | Authors: | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | Deposit date: | 2008-06-09 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
3DD5
 
 | | Glomerella cingulata E600-cutinase complex | | Descriptor: | Cutinase, DIETHYL PHOSPHONATE | | Authors: | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | Deposit date: | 2008-06-05 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
3BZQ
 
 | | High resolution crystal structure of Nitrogen Regulatory Protein (Rv2919c) of Mycobacterium tuberculosis | | Descriptor: | Nitrogen regulatory protein P-II | | Authors: | Reddy, M.C, Palaninathan, S.K, Shetty, N.D, Owen, J.L, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of the apo and ATP bound Mycobacterium tuberculosis nitrogen regulatory PII protein.
Protein Sci., 19, 2010
|
|
2A97
 
 | |
4M1A
 
 | | Crystal structure of a Domain of unknown function (DUF1904) from Sebaldella termitidis ATCC 33386 | | Descriptor: | Hypothetical protein | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-02 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a Domain of unknown function (DUF1904) from Sebaldellatermitidis ATCC 33386
To be Published
|
|
