2LOH
 
 | |
2LLM
 
 | |
6HJ7
 
 | |
7S8B
 
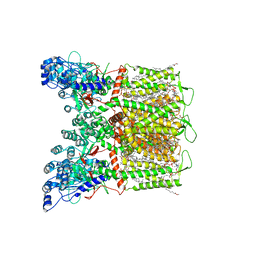 | | Cryo-EM structure of human TRPV6 in complex with channel blocker ruthenium red | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2021-09-17 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Structural mechanisms of TRPV6 inhibition by ruthenium red and econazole.
Nat Commun, 12, 2021
|
|
6F61
 
 | |
2KGU
 
 | | Spatial structure of purotoxin-1 in water | | Descriptor: | Purotoxin-1 | | Authors: | Nadezhdin, K, Vassilevski, A, Arseniev, A, Grishin, E, Pluzhnikov, K, Korolkova, Y. | | Deposit date: | 2009-03-20 | | Release date: | 2010-03-31 | | Last modified: | 2019-05-15 | | Method: | SOLUTION NMR | | Cite: | Novel peptide from spider venom inhibits P2X3 receptors and inflammatory pain.
Ann Neurol, 67, 2010
|
|
6RRO
 
 | |
6RRL
 
 | |
2MJO
 
 | |
2MIC
 
 | |
2N90
 
 | |
8W2L
 
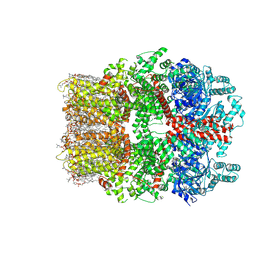 | | TRPM7 structure in complex with anticancer agent CCT128930 in closed state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 4-(4-chlorobenzyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-aminium, ... | | Authors: | Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2024-02-20 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural basis of selective TRPM7 inhibition by the anticancer agent CCT128930.
Cell Rep, 43, 2024
|
|
2JWM
 
 | |
9B37
 
 | | Open state of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to one concanavalin A dimer. Composite map. | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nadezhdin, K.D, Gangwar, S.P, Sobolevsky, A.I. | | Deposit date: | 2024-03-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (6.66 Å) | | Cite: | Kainate receptor channel opening and gating mechanism.
Nature, 630, 2024
|
|
9B38
 
 | | Kainate receptor GluK2 in complex with agonist glutamate with pseudo 4-fold symmetrical ligand-binding domain layer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Nadezhdin, K.D, Gangwar, S.P, Sobolevsky, A.I. | | Deposit date: | 2024-03-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Kainate receptor channel opening and gating mechanism.
Nature, 630, 2024
|
|
9B39
 
 | | Kainate receptor GluK2 in complex with agonist glutamate with asymmetric ligand-binding domain layer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Nadezhdin, K.D, Gangwar, S.P, Sobolevsky, A.I. | | Deposit date: | 2024-03-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Kainate receptor channel opening and gating mechanism.
Nature, 630, 2024
|
|
9B36
 
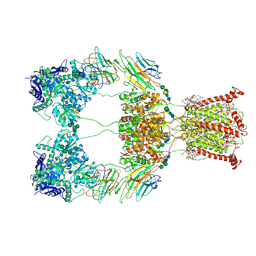 | | Open state of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to two concanavalin A dimers. Composite map. | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nadezhdin, K.D, Gangwar, S.P, Sobolevsky, A.I. | | Deposit date: | 2024-03-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Kainate receptor channel opening and gating mechanism.
Nature, 630, 2024
|
|
2MZG
 
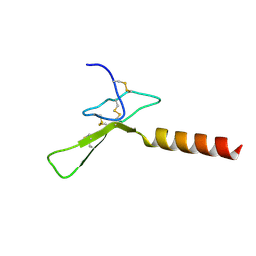 | | Purotoxin-2 NMR structure in DPC micelles | | Descriptor: | Purotoxin-2 | | Authors: | Nadezhdin, K, Oparin, P, Vassilevski, A, Grishin, E, Arseniev, A. | | Deposit date: | 2015-02-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of purotoxin-2 from wolf spider: modular design and membrane-assisted mode of action in arachnid toxins.
Biochem. J., 473, 2016
|
|
2N86
 
 | | NMR structure of OtTx1a - ICK | | Descriptor: | Spiderine-1a | | Authors: | Nadezhdin, K, Romanovskaya, D, Sachkova, M, Vassilevski, A, Grishin, E, Kovalchuk, S, Arseniev, A. | | Deposit date: | 2015-10-05 | | Release date: | 2016-10-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Modular toxin from the lynx spider Oxyopes takobius: Structure of spiderine domains in solution and membrane-mimicking environment.
Protein Sci., 26, 2017
|
|
2MZF
 
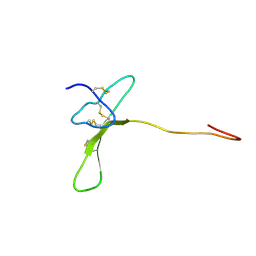 | | Purotoxin-2 NMR structure in water | | Descriptor: | Purotoxin-2 | | Authors: | Nadezhdin, K, Vassilevski, A, Oparin, P, Grishin, E, Arseniev, A. | | Deposit date: | 2015-02-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of purotoxin-2 from wolf spider: modular design and membrane-assisted mode of action in arachnid toxins.
Biochem. J., 473, 2016
|
|
2N85
 
 | | NMR structure of OtTx1a - AMP in DPC micelles | | Descriptor: | Spiderine-1a | | Authors: | Nadezhdin, K, Romanovskaya, D, Sachkova, M, Vassilevski, A, Grishin, E, Kovalchuk, S, Arseniev, A. | | Deposit date: | 2015-10-05 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Modular toxin from the lynx spider Oxyopes takobius: Structure of spiderine domains in solution and membrane-mimicking environment.
Protein Sci., 26, 2017
|
|
9B35
 
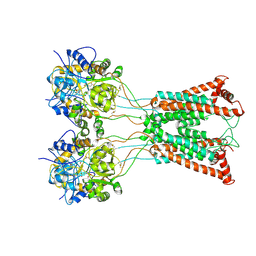 | | Ligand-binding and transmembrane domains of kainate receptor GluK2 in the open state, a complex with agonist glutamate and positive allosteric modulator BPAM344 | | Descriptor: | GLUTAMIC ACID, Glutamate receptor ionotropic, kainate 2 | | Authors: | Nadezhdin, K.D, Gangwar, S.P, Sobolevsky, A.I. | | Deposit date: | 2024-03-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Kainate receptor channel opening and gating mechanism.
Nature, 630, 2024
|
|
9B33
 
 | |
9B34
 
 | |
7LQY
 
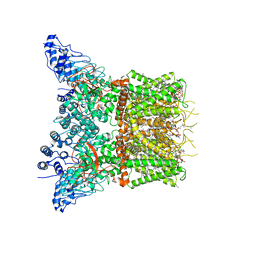 | | Structure of squirrel TRPV1 in apo state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoinositol, CHLORIDE ION, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2021-02-15 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Extracellular cap domain is an essential component of the TRPV1 gating mechanism.
Nat Commun, 12, 2021
|
|
