2YTT
 
 | | Solution structure of the C2H2 type zinc finger (region 204-236) of human Zinc finger protein 473 | | Descriptor: | ZINC ION, Zinc finger protein 473 | | Authors: | Tochio, N, Tomizawa, T, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 204-236) of human Zinc finger protein 473
To be Published
|
|
8RCW
 
 | | Crystal structure of the Mycobacterium tuberculosis regulator VirS (N-terminal fragment 4-208) in complex with the lead compound SMARt751 | | Descriptor: | 4,4,4-tris(fluoranyl)-1-[4-(4-fluorophenyl)piperidin-1-yl]butan-1-one, HTH-type transcriptional regulator VirS | | Authors: | Grosse, C, Sigoillot, M, Megalizzi, V, Tanina, A, Willand, N, Baulard, A.R, Wintjens, R. | | Deposit date: | 2023-12-07 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis VirS regulator reveals its interaction with the lead compound SMARt751.
J.Struct.Biol., 216, 2024
|
|
6F5C
 
 | |
8QWR
 
 | | Crystal structure of CotB2 variant V80L | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Cyclooctat-9-en-7-ol synthase, ... | | Authors: | Dimos, N, Himpich, S, Ringel, M, Driller, R, Major, D.T, Brueck, T, Loll, B. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of CotB2 variant V80L
To Be Published
|
|
8IPG
 
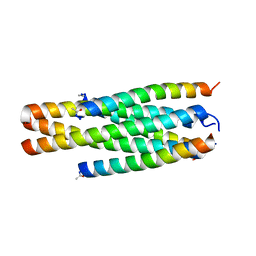 | | Structure of HP101/N44 | | Descriptor: | Env polyprotein (Fragment), HP101 | | Authors: | Liu, N, Qin, B. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of HP101/N44
To Be Published
|
|
7Z0J
 
 | | human PEX13 SH3 domain in complex with internal FxxxF motif | | Descriptor: | 1,2-ETHANEDIOL, Peroxisomal membrane protein PEX13 | | Authors: | Gaussmann, S, Zak, K, Kreisz, N, Sattler, M. | | Deposit date: | 2022-02-23 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Intramolecular autoinhibition of human PEX13 modulates peroxisomal import
Biorxiv, 2022
|
|
7Q11
 
 | | Crystal structure of CTX-M-14 in complex with Ixazomib | | Descriptor: | Beta-lactamase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Werner, N, Perbandt, M, Hinrichs, W, Prester, A, Rohde, H, Aepfelbacher, M, Betzel, C. | | Deposit date: | 2021-10-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structural basis to repurpose boron-based proteasome inhibitors Bortezomib and Ixazomib as beta-lactamase inhibitors.
Sci Rep, 12, 2022
|
|
8QWS
 
 | | Crystal structure of CotB2 variant V80L in complex with alendronate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Dimos, N, Himpich, S, Ringel, M, Driller, R, Major, D.T, Brueck, T, Loll, B. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of CotB2 variant V80L in complex with alendronate
To Be Published
|
|
7Q0Y
 
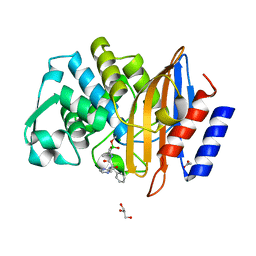 | | Crystal structure of CTX-M-14 in complex with Bortezomib | | Descriptor: | ACETATE ION, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Werner, N, Perbandt, M, Hinrichs, W, Prester, A, Rohde, H, Aepfelbacher, M, Betzel, C. | | Deposit date: | 2021-10-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis to repurpose boron-based proteasome inhibitors Bortezomib and Ixazomib as beta-lactamase inhibitors.
Sci Rep, 12, 2022
|
|
7Q0Z
 
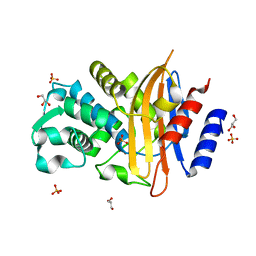 | | Crystal structure of CTX-M-14 | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Werner, N, Perbandt, M, Hinrichs, W, Prester, A, Rohde, H, Aepfelbacher, M, Betzel, C. | | Deposit date: | 2021-10-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural basis to repurpose boron-based proteasome inhibitors Bortezomib and Ixazomib as beta-lactamase inhibitors.
Sci Rep, 12, 2022
|
|
4XU5
 
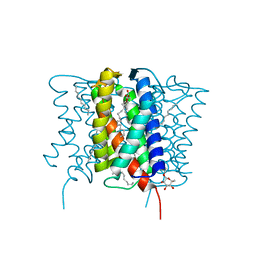 | | Crystal structure of MvINS bound to a bromine-derived 14C Diacylglycerol (DAG) at 2.1A resolution | | Descriptor: | (2S)-1-[(13-bromotridecanoyl)oxy]-3-hydroxypropan-2-yl tetradecanoate, DECANE, Uncharacterized protein, ... | | Authors: | Ren, R.B, Wu, J.P, Yan, C.Y, He, Y, Yan, N. | | Deposit date: | 2015-01-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels
Science, 349, 2015
|
|
6EYJ
 
 | | E-selectin lectin, EGF-like and two SCR domains complexed with glycomimetic ligand NV354 | | Descriptor: | (2~{S})-3-cyclohexyl-2-[(2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-3,5-bis(oxidanyl)-6-[(1~{R},2~{R})-2-[(2~{R},3~{S},4~{R},5~{S},6~{R})-3,4,5-tris(oxidanyl)-6-(trifluoromethyl)oxan-2-yl]oxycyclohexyl]oxy-oxan-4-yl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jakob, R.P, Zihlmann, P, Preston, R.C, Varga, N, Ernst, B, Maier, T. | | Deposit date: | 2017-11-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | E-selectin lectin with different glycomimetic ligands
To Be Published
|
|
6EZ1
 
 | |
4XAF
 
 | | Cycles of destabilization and repair underlie evolutionary transitions in enzymes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R1, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-14 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
7Z2K
 
 | | Crystal structure of SARS-CoV-2 Main Protease in orthorhombic space group p212121 | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2022-02-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
8HU1
 
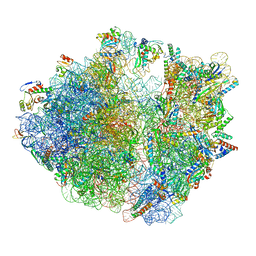 | | E. coli 70S ribosome complexed with tRNA_Ile2 bearing L34 and ct6A37 in classical state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-12-22 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural insights into the decoding capability of isoleucine tRNAs with lysidine and agmatidine.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HTZ
 
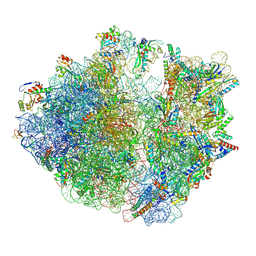 | | E. coli 70S ribosome complexed with H. marismortui tRNA_Ile2 bearing agm2C34 in classical state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-12-22 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural insights into the decoding capability of isoleucine tRNAs with lysidine and agmatidine.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6F5H
 
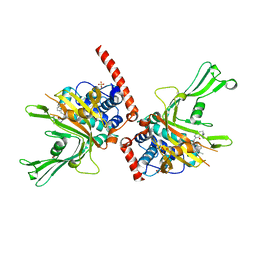 | | Crystal structure of USP7 in complex with a 4-hydroxypiperidine based inhibitor | | Descriptor: | 3-[[4-oxidanyl-1-[(3~{R})-3-phenylbutanoyl]piperidin-4-yl]methyl]-6-(2-pyrrolidin-1-ylethylamino)pyrimidin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Harrison, T, Gavory, G, O'Dowd, C, Helm, M, Flasz, J, Dossang, A, Hughes, C, Cassidy, E, McClelland, K, Odrzywol, E, Page, N, Barker, O, Miel, H, Feutron-Burton, S, Rountree, J.S.S. | | Deposit date: | 2017-12-01 | | Release date: | 2018-04-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Identification and Structure-Guided Development of Pyrimidinone Based USP7 Inhibitors.
ACS Med Chem Lett, 9, 2018
|
|
8QQM
 
 | | nicotinic acetylcholine receptor in intact synaptic membrane | | Descriptor: | Acetylcholine receptor subunit alpha, Acetylcholine receptor subunit beta, Acetylcholine receptor subunit delta, ... | | Authors: | Unwin, N. | | Deposit date: | 2023-10-05 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Influence of lipid bilayer on the structure of the muscle-type nicotinic acetylcholine receptor.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7PYV
 
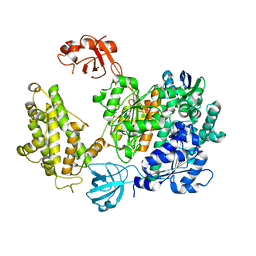 | | Crystal structure of human UBA6 in complex with the ubiquitin-like modifier FAT10 | | Descriptor: | UBD, Ubiquitin-like modifier-activating enzyme 6,Ubiquitin-like modifier-activating enzyme 1,Ubiquitin-like modifier-activating enzyme 6 | | Authors: | Li, S, Truongvan, N, Schindelin, H. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
8I41
 
 | |
8I42
 
 | |
8QAM
 
 | |
8I48
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in closed state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | Authors: | Bharambe, N, Li, Z, Basak, S. | | Deposit date: | 2023-01-18 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
1WFS
 
 | | Solution Structure of Glia Maturation Factor-gamma from Mus Musculus | | Descriptor: | Glia maturation factor gamma | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-26 | | Release date: | 2004-11-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains.
Protein Sci., 18, 2009
|
|
