5CU8
 
 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 2-Amino-6-chlorobenzothiazole (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 6-chloro-1,3-benzothiazol-2-amine, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 2-Amino-6-chlorobenzothiazole (SGC - Diamond I04-1 fragment screening)
To be published
|
|
1RYU
 
 | | Solution Structure of the SWI1 ARID | | Descriptor: | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin subfamily F member 1 | | Authors: | Kim, S, Zhang, Z, Upchurch, S, Isern, N, Chen, Y. | | Deposit date: | 2003-12-22 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and DNA-binding sites of the SWI1 AT-rich interaction domain (ARID) suggest determinants for sequence-specific DNA recognition.
J.Biol.Chem., 279, 2004
|
|
5CUE
 
 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with AGN-PC-04G0SS (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-N,1-dimethyl-1H-pyrazole-4-carboxamide, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with AGN-PC-04G0SS (SGC - Diamond I04-1 fragment screening)
To be published
|
|
1S5Z
 
 | | NDP kinase in complex with adenosine phosphonoacetic acid | | Descriptor: | ADENOSINE PHOSPHONOACETIC ACID, Nucleoside diphosphate kinase, cytosolic, ... | | Authors: | Chen, Y, Morera, S, Pasti, C, Angusti, A, Solaroli, N, Veron, M, Janin, J, Manfredini, S, Deville-Bonne, D. | | Deposit date: | 2004-01-22 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Adenosine phosphonoacetic acid is slowly metabolized by NDP kinase.
Med.Chem., 1, 2005
|
|
8B55
 
 | | Human ADGRG4 PTX-like domain | | Descriptor: | Adhesion G-protein coupled receptor G4, MAGNESIUM ION | | Authors: | Kieslich, B, Straeter, N. | | Deposit date: | 2022-09-21 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The dimerized pentraxin-like domain of the adhesion G protein-coupled receptor 112 (ADGRG4) suggests function in sensing mechanical forces.
J.Biol.Chem., 299, 2023
|
|
6C8S
 
 | | Loganic acid methyltransferase with SAH | | Descriptor: | Loganic acid O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Petronikolou, N, Nair, S.K. | | Deposit date: | 2018-01-25 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Loganic Acid Methyltransferase: Insights into the Specificity of Methylation on an Iridoid Glycoside.
Chembiochem, 19, 2018
|
|
1S23
 
 | | Crystal Structure Analysis of the B-DNA Decamer CGCAATTGCG | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*TP*TP*GP*CP*G)-3', COBALT (II) ION | | Authors: | Valls, N, Wright, G, Steiner, R.A, Murshudov, G.N, Subirana, J.A. | | Deposit date: | 2004-01-08 | | Release date: | 2004-04-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA variability in five crystal structures of d(CGCAATTGCG).
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6CAA
 
 | | CryoEM structure of human SLC4A4 sodium-coupled acid-base transporter NBCe1 | | Descriptor: | Electrogenic sodium bicarbonate cotransporter 1 | | Authors: | Huynh, K.W, Jiang, J, Abuladze, N, Tsirulnikov, K, Kao, L, Shao, X, Newman, D, Azimov, R, Pushkin, A, Zhou, Z.H, Kurtz, I. | | Deposit date: | 2018-01-29 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | CryoEM structure of the human SLC4A4 sodium-coupled acid-base transporter NBCe1.
Nat Commun, 9, 2018
|
|
6W5U
 
 | | NPC1 structure in GDN micelles at pH 5.5, conformation b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Yan, N, Qian, H.W, Wu, X.L. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6C1K
 
 | | HypoPP mutant with ligand1 | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GUANIDINE, Ion transport protein, ... | | Authors: | Catterall, W.A, Zheng, N, Jiang, D, Gamal El-Din, T.M. | | Deposit date: | 2018-01-04 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for gating pore current in periodic paralysis.
Nature, 557, 2018
|
|
6GQ1
 
 | | Cryo-EM reconstruction of yeast 80S ribosome in complex with mRNA, tRNA and eEF2 (GMPPCP/sordarin) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Pellegrino, S, Demeshkina, N, Mancera-Martinez, E, Melnikov, S, Simonetti, A, Myasnikov, A, Yusupov, M, Yusupova, G, Hashem, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-07-11 | | Last modified: | 2018-08-08 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Insights into the Role of Diphthamide on Elongation Factor 2 in mRNA Reading-Frame Maintenance.
J. Mol. Biol., 430, 2018
|
|
6QAI
 
 | | 2-desoxiribosyltransferase from Leishmania mexicana | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-Deoxyribosyltransferase | | Authors: | Crespo, N, Mancheno, J.M. | | Deposit date: | 2018-12-19 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | 2'-Deoxyribosyltransferase from Leishmania mexicana, an efficient biocatalyst for one-pot, one-step synthesis of nucleosides from poorly soluble purine bases.
Appl.Microbiol.Biotechnol., 101, 2017
|
|
6QBJ
 
 | |
6GVH
 
 | | Crystal structure of PI3K alpha in complex with 3-(2-Amino-benzooxazol-5-yl)-4-chloro-1-isopropyl-1H-pyrazolo[3,4-d]pyrimidin-6-ylamine | | Descriptor: | 5-(6-azanyl-4-chloranyl-1-propan-2-yl-pyrazolo[3,4-d]pyrimidin-3-yl)-1,3-benzoxazol-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Ouvry, G, Aurelly, M, Bonnary, L, Borde, E, Bouix-Peter, C, Chantalat, L, Clary, L, Defoin-Platel, C, Deret, S, Forissier, M, Harris, C.S, Isabet, T, Lamy, L, Luzy, A.P, Pascau, J, Soulet, C, Taddei, A, Taquet, N, Tomas, L, Thoreau, E, Varvier, E, Vial, E, Hennequin, L.F. | | Deposit date: | 2018-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Impact of Minor Structural Modifications on Properties of a Series of mTOR Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
5D9B
 
 | | Luciferin-regenerating enzyme solved by SIRAS using XFEL (refined against native data) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography.
Sci Rep, 5, 2015
|
|
6C8F
 
 | | Crystal structure of Transient Receptor Potential (TRP) channel TRPV4 in the presence of cesium | | Descriptor: | CESIUM ION, Transient receptor potential cation channel, subfamily V, ... | | Authors: | Deng, Z, Paknejad, N, Maksaev, G, Sala-Rabanal, M, Nichols, C.G, Hite, R.K, Yuan, P. | | Deposit date: | 2018-01-24 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6W6O
 
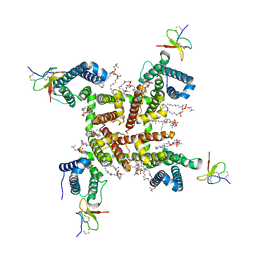 | | NaChBac-Nav1.7VSDII chimera and HWTX-IV complex | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Huwentoxin-IV, NaChBac-Nav1.7VSDII chimera | | Authors: | Yan, N, Gao, S. | | Deposit date: | 2020-03-17 | | Release date: | 2020-06-24 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Employing NaChBac for cryo-EM analysis of toxin action on voltage-gated Na + channels in nanodisc.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6GWS
 
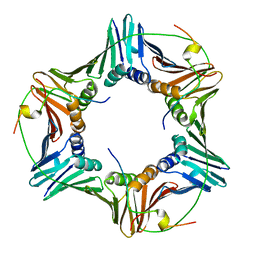 | | Crystal structure of human PCNA in complex with three p15 peptides | | Descriptor: | PCNA-associated factor, Proliferating cell nuclear antigen | | Authors: | De March, M, Merino, N, Gonzalez-Magana, A, Romano-Moreno, M, Onesti, S, Blanco, F.J, De Biasio, A. | | Deposit date: | 2018-06-25 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | p15PAF binding to PCNA modulates the DNA sliding surface.
Nucleic Acids Res., 46, 2018
|
|
6GJC
 
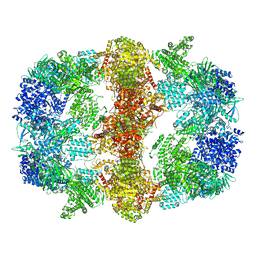 | | Structure of Mycobacterium tuberculosis Fatty Acid Synthase - I | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase | | Authors: | Elad, N, Baron, S, Shakked, Z, Zimhony, O, Diskin, R. | | Deposit date: | 2018-05-16 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of Type-I Mycobacterium tuberculosis fatty acid synthase at 3.3 angstrom resolution.
Nat Commun, 9, 2018
|
|
1SQZ
 
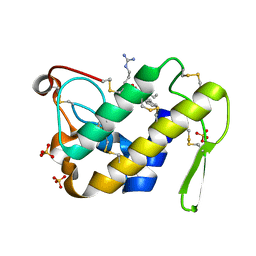 | | Design of specific inhibitors of Phopholipase A2: Crystal structure of the complex formed between Group II Phopholipase A2 and a designed peptide Dehydro-Ile-Ala-Arg-Ser at 1.2A resolution | | Descriptor: | Phospholipase A2, SULFATE ION, synthetic peptide | | Authors: | Singh, N, Prem Kumar, R, Somvanshi, R.K, Bilgrami, S, Ethayathulla, A.S, Sharma, S, Dey, S, Singh, T.P. | | Deposit date: | 2004-03-22 | | Release date: | 2004-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Design of specific inhibitors of Phopholipase A2: Crystal structure of the complex formed between GroupII Phopholipase A2 and a designed peptide Dehydro-Ile-Ala-Arg-Ser at 1.2A resolution
To be Published
|
|
6VZR
 
 | | Engineered TTLL6 bound to the initiation analog | | Descriptor: | (2~{S})-2-[[[(3~{R})-3-acetamido-4-(ethylamino)-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6GSF
 
 | | Solution structure of lipase binding domain LID1 of foldase from Pseudomonas aeruginosa | | Descriptor: | Lipase chaperone | | Authors: | Viegas, A, Jaeger, K.-E, Etzkorn, M, Gohlke, H, Verma, N, Dollinger, P, Kovacic, F. | | Deposit date: | 2018-06-14 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic insights revealing how lipase binding domain MD1 of Pseudomonas aeruginosa foldase affects lipase activation.
Sci Rep, 10, 2020
|
|
5D8W
 
 | | Structrue of a lucidum protein | | Descriptor: | Endoglucanase | | Authors: | Guo, R, Li, Q, Shang, N, Liu, G, Ko, T.P, Chen, C.C, Liu, W. | | Deposit date: | 2015-08-18 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Functional and structural analyses of a 1,4-beta-endoglucanase from Ganoderma lucidum.
Enzyme.Microb.Technol., 86, 2016
|
|
5D98
 
 | | Influenza C Virus RNA-dependent RNA Polymerase - Space group P43212 | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Hengrung, N, El Omari, K, Serna Martin, I, Vreede, F.T, Cusack, S, Rambo, R.P, Vonrhein, C, Bricogne, G, Stuart, D.I, Grimes, J.M, Fodor, E. | | Deposit date: | 2015-08-18 | | Release date: | 2015-10-21 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase from influenza C virus.
Nature, 527, 2015
|
|
6BV8
 
 | | Structure of proteinaceous RNase P 1 (PRORP1) from A. thaliana complexed with Mn after 3-hour soak with juglone | | Descriptor: | 5-hydroxynaphthalene-1,4-dione, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Karasik, A, Wu, N, Fierke, C.A, Koutmos, M. | | Deposit date: | 2017-12-12 | | Release date: | 2019-06-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of protein-only RNase P with Gambogic acid and Juglone
To Be Published
|
|
