7PBY
 
 | |
6WO3
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 6a bound to broadly neutralizing antibody U1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2020-04-24 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | An alternate conformation of HCV E2 neutralizing face as an additional vaccine target.
Sci Adv, 6, 2020
|
|
7PJT
 
 | | Structure of the 70S ribosome with tRNAs in hybrid state 1 (H1) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petrychenko, V, Peng, B.Z, Schwarzer, A.C, Peske, F, Rodnina, M.V, Fischer, N. | | Deposit date: | 2021-08-24 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural mechanism of GTPase-powered ribosome-tRNA movement.
Nat Commun, 12, 2021
|
|
7PJX
 
 | | Structure of the 70S-EF-G-GDP ribosome complex with tRNAs in hybrid state 1 (H1-EF-G-GDP) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petrychenko, V, Peng, B.Z, Schwarzer, A.C, Peske, F, Rodnina, M.V, Fischer, N. | | Deposit date: | 2021-08-24 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structural mechanism of GTPase-powered ribosome-tRNA movement.
Nat Commun, 12, 2021
|
|
7PJW
 
 | | Structure of the 70S-EF-G-GDP-Pi ribosome complex with tRNAs in hybrid state 2 (H2-EF-G-GDP-Pi) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petrychenko, V, Peng, B.Z, Schwarzer, A.C, Peske, F, Rodnina, M.V, Fischer, N. | | Deposit date: | 2021-08-24 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural mechanism of GTPase-powered ribosome-tRNA movement.
Nat Commun, 12, 2021
|
|
6W5V
 
 | | NPC1-NPC2 complex structure at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Yan, N, Qian, H.W, Wu, X.L. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
7PJY
 
 | | Structure of the 70S-EF-G-GDP ribosome complex with tRNAs in chimeric state 1 (CHI1-EF-G-GDP) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petrychenko, V, Peng, B.Z, Schwarzer, A.C, Peske, F, Rodnina, M.V, Fischer, N. | | Deposit date: | 2021-08-24 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural mechanism of GTPase-powered ribosome-tRNA movement.
Nat Commun, 12, 2021
|
|
6VJ7
 
 | | Crystal structure of red kidney bean purple acid phosphatase in complex with adenosine 5'-(beta,gamma imido)triphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, Schenk, G, Guddat, L.W, McGeary, R.P, Mitic, N, Furtado, A, Schulz, B.L, Henry, R.J, Schmidt, S. | | Deposit date: | 2020-01-15 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural elements that modulate the substrate specificity of plant purple acid phosphatases: Avenues for improved phosphorus acquisition in crops.
Plant Sci., 294, 2020
|
|
6VRC
 
 | | Cryo-EM structure of Cas13(crRNA) | | Descriptor: | CRISPR-associated endoribonuclease Cas13a, RNA (51-MER) | | Authors: | Jia, N, Meeske, A.J, Marraffini, L.A, Patel, D.J. | | Deposit date: | 2020-02-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity.
Science, 369, 2020
|
|
8FLI
 
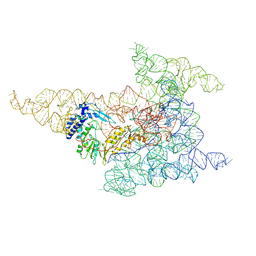 | | Cryo-EM structure of a group II intron immediately before branching | | Descriptor: | Group II Intron, MAGNESIUM ION, Maturase reverse transcriptase | | Authors: | Haack, D.B, Rudolfs, B.G, Zhang, C, Lyumkis, D, Toor, N. | | Deposit date: | 2022-12-21 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of branching during RNA splicing.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6VYH
 
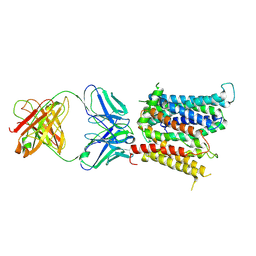 | | Cryo-EM structure of SLC40/ferroportin in complex with Fab | | Descriptor: | 11F9 heavy-chain, 11F9 light-chain, COBALT (II) ION, ... | | Authors: | Shen, J, Ren, Z, Pan, Y, Gao, S, Yan, N, Zhou, M. | | Deposit date: | 2020-02-26 | | Release date: | 2020-11-11 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of ion transport and inhibition in ferroportin.
Nat Commun, 11, 2020
|
|
6WYC
 
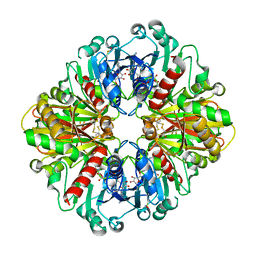 | |
6WCC
 
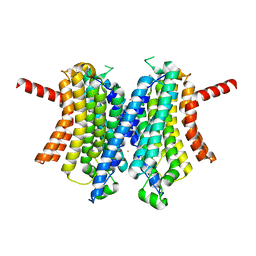 | | Human closed state TMEM175 in CsCl | | Descriptor: | CESIUM ION, Endosomal/lysosomal potassium channel TMEM175 | | Authors: | Oh, S, Paknejad, N, Hite, R.K. | | Deposit date: | 2020-03-30 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Gating and selectivity mechanisms for the lysosomal K + channel TMEM175.
Elife, 9, 2020
|
|
6WHT
 
 | | GluN1b-GluN2B NMDA receptor in active conformation at 4.4 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.39 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6WIK
 
 | | Cryo-EM structure of SLC40/ferroportin with Fab in the presence of hepcidin | | Descriptor: | 11F9 Fab heavy-chain, 11F9 Fab light-chain, Solute carrier family 40 protein | | Authors: | Shen, J, Ren, Z, Pan, Y, Gao, S, Yan, N, Zhou, M. | | Deposit date: | 2020-04-10 | | Release date: | 2020-11-11 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of ion transport and inhibition in ferroportin.
Nat Commun, 11, 2020
|
|
8EZL
 
 | | Pfs25 in complex with transmission-reducing antibody AS01-50 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 25 kDa ookinete surface antigen, Transmission-reducing antibody AS01-50, ... | | Authors: | Shukla, N, Tang, W.K, Tolia, N.H. | | Deposit date: | 2022-11-01 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A human antibody epitope map of the malaria vaccine antigen Pfs25.
Npj Vaccines, 8, 2023
|
|
6WI0
 
 | | GluN1b-GluN2B NMDA receptor in complex with GluN1 antagonist L689,560, class 2 | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-15 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6WOQ
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 1a bound to neutralizing antibody HC1AM and non neutralizing antibody E1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2020-04-25 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.667 Å) | | Cite: | An alternate conformation of HCV E2 neutralizing face as an additional vaccine target.
Sci Adv, 6, 2020
|
|
6WPF
 
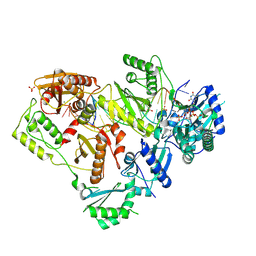 | | Structure of HIV-1 Reverse Transcriptase (RT) in complex with dsDNA and d4T | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA Primer 21-mer, DNA template 27-mer, ... | | Authors: | Bertoletti, N, Anderson, K.S. | | Deposit date: | 2020-04-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Post-Catalytic Complexes with Emtricitabine or Stavudine and HIV-1 Reverse Transcriptase Reveal New Mechanistic Insights for Nucleotide Incorporation and Drug Resistance.
Molecules, 25, 2020
|
|
6WV6
 
 | | Human VKOR with phenindione | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Phenindione, Vitamin K epoxide reductase, ... | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
7QDF
 
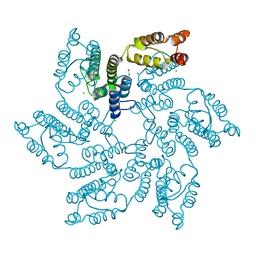 | | Hexameric HIV-1 (M-group) CA R120 mutant | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Gag polyprotein, ... | | Authors: | Govasli, M.A.L, Pinotsis, N, McAlpine-Scott, S. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Evasion of cGAS and TRIM5 defines pandemic HIV.
Nat Microbiol, 7, 2022
|
|
8FHS
 
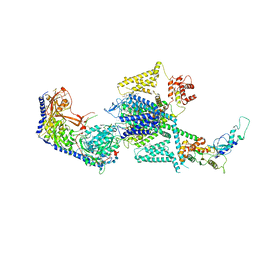 | | Human L-type voltage-gated calcium channel Cav1.2 in the presence of amiodarone and sofosbuvir at 3.3 Angstrom resolution | | Descriptor: | (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
6WI1
 
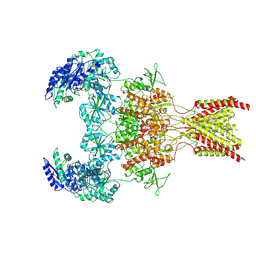 | | GluN1b-GluN2B NMDA receptor in active conformation stabilized by inter-GluN1b-GluN2B subunit cross-linking | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ionotropic glutamate receptor , ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6WO4
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 6a bound to broadly neutralizing antibody HC11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2020-04-24 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | An alternate conformation of HCV E2 neutralizing face as an additional vaccine target.
Sci Adv, 6, 2020
|
|
8FOR
 
 | |
