1B4K
 
 | | High resolution crystal structure of a MG2-dependent 5-aminolevulinic acid dehydratase | | Descriptor: | LAEVULINIC ACID, MAGNESIUM ION, PROTEIN (5-AMINOLEVULINIC ACID DEHYDRATASE), ... | | Authors: | Frankenberg, N, Jahn, D, Heinz, D.W. | | Deposit date: | 1998-12-22 | | Release date: | 1999-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | High resolution crystal structure of a Mg2+-dependent porphobilinogen synthase.
J.Mol.Biol., 289, 1999
|
|
5FL1
 
 | | Structure of a hydrolase with an inhibitor | | Descriptor: | (3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-5-(hydroxymethyl)-2-(prop-2-enylamino)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]thiazole-6,7-diol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
5FO2
 
 | | Structure of human transthyretin mutant A108I | | Descriptor: | TRANSTHYRETIN | | Authors: | Varejao, N, Santanna, R, Saraiva, M.J, Gallego, P, Ventura, S, Reverter, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Cavity filling mutations at the thyroxine-binding site dramatically increase transthyretin stability and prevent its aggregation.
Sci Rep, 7, 2017
|
|
1BKE
 
 | | HUMAN SERUM ALBUMIN IN A COMPLEX WITH MYRISTIC ACID AND TRI-IODOBENZOIC ACID | | Descriptor: | 2,3,5-TRIIODOBENZOIC ACID, MYRISTIC ACID, SERUM ALBUMIN | | Authors: | Curry, S, Mandelkow, H, Brick, P, Franks, N. | | Deposit date: | 1998-07-06 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites.
Nat.Struct.Biol., 5, 1998
|
|
1TG1
 
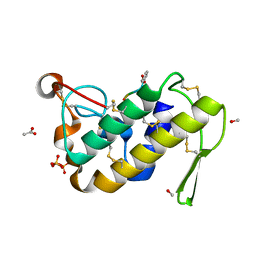 | | Crystal Structure of the complex formed between russells viper phospholipase A2 and a designed peptide inhibitor PHQ-Leu-Val-Arg-Tyr at 1.2A resolution | | Descriptor: | ACETIC ACID, METHANOL, Phospholipase A2, ... | | Authors: | Singh, N, Kaur, P, Somvanshi, R.K, Sharma, S, Dey, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2004-05-28 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of the complex formed between russells viper phospholipase A2 and a designed peptide inhibitor Cbz-dehydro-Leu-Val-Arg-Tyr at 1.2A resolution
To be Published
|
|
5FAR
 
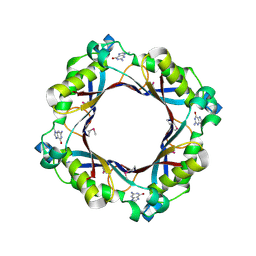 | | Crystal structure of dihydroneopterin aldolase from Bacillus anthracis complex with 9-METHYLGUANINE | | Descriptor: | 7,8-dihydroneopterin aldolase, 9-METHYLGUANINE | | Authors: | Chang, C, Maltseva, N, Kim, Y, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of dihydroneopterin aldolase from Bacillus anthracis complex with 9-METHYLGUANINE
To Be Published
|
|
7FBR
 
 | |
5FBM
 
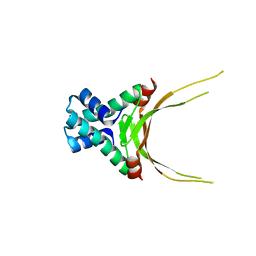 | | Crystal Structure of Histone Like Protein (HLP) from Streptococcus mutans Refined to 1.9 A Resolution | | Descriptor: | DNA-binding protein HU | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, O'Neil, P, Biswas, I. | | Deposit date: | 2015-12-14 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of histone-like protein from Streptococcus mutans refined to 1.9 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
1BZE
 
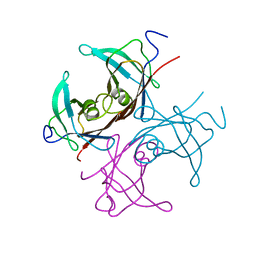 | |
1C0F
 
 | | CRYSTAL STRUCTURE OF DICTYOSTELIUM CAATP-ACTIN IN COMPLEX WITH GELSOLIN SEGMENT 1 | | Descriptor: | ACTIN, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Matsuura, Y, Stewart, M, Kawamoto, M, Kamiya, N, Saeki, K, Yasunaga, T, Wakabayashi, T. | | Deposit date: | 1999-07-16 | | Release date: | 2000-03-01 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the higher Ca(2+)-activation of the regulated actin-activated myosin ATPase observed with Dictyostelium/Tetrahymena actin chimeras
J.Mol.Biol., 296, 2000
|
|
5FZ7
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with Maybridge fragment ethyl 2-amino-4-thiophen-2-ylthiophene-3- carboxylate (N06131b) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Kupinska, K, Szykowska, A, Pearce, N, Talon, R, Collins, P, Gileadi, C, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-11 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Maybridge Fragment Ethyl 2-Amino-4-Thiophen-2-Ylthiophene-3-Carboxylate (N06131B) (Ligand Modelled Based on Pandda Event Map, Sgc - Diamond I04-1 Fragment Screening)
To be Published
|
|
7Z1U
 
 | | Biochemical implications of the substitution of a unique cysteine residue in sugar beet phytoglobin BvPgb 1.2 | | Descriptor: | Non-symbiotic hemoglobin class 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nyblom, M, Christensen, S, Leiva Eriksson, N, Bulow, L. | | Deposit date: | 2022-02-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Oxidative Implications of Substituting a Conserved Cysteine Residue in Sugar Beet Phytoglobin BvPgb 1.2.
Antioxidants, 11, 2022
|
|
2WSI
 
 | |
2WZX
 
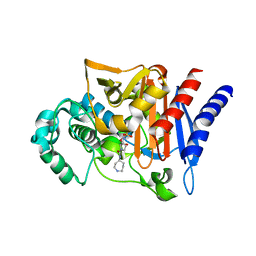 | |
1BRE
 
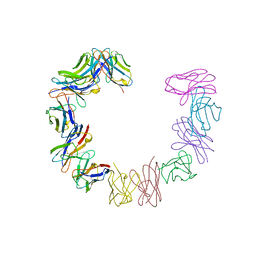 | | IMMUNOGLOBULIN LIGHT CHAIN PROTEIN | | Descriptor: | BENCE-JONES KAPPA I PROTEIN BRE | | Authors: | Schormann, N, Benson, M.D. | | Deposit date: | 1995-07-19 | | Release date: | 1995-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tertiary structure of an amyloid immunoglobulin light chain protein: a proposed model for amyloid fibril formation.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1BWV
 
 | | Activated Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase (RUBISCO) Complexed with the Reaction Intermediate Analogue 2-Carboxyarabinitol 1,5-Bisphosphate | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (RIBULOSE BISPHOSPHATE CARBOXYLASE) | | Authors: | Sugawara, H, Yamamoto, H, Shibata, N, Inoue, T, Miyake, C, Yokota, A, Kai, Y. | | Deposit date: | 1998-09-29 | | Release date: | 1999-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of carboxylase reaction-oriented ribulose 1, 5-bisphosphate carboxylase/oxygenase from a thermophilic red alga, Galdieria partita.
J.Biol.Chem., 274, 1999
|
|
1PCN
 
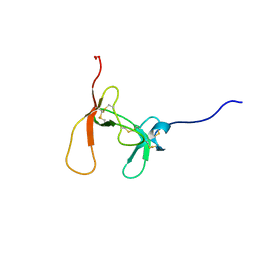 | | SOLUTION STRUCTURE OF PORCINE PANCREATIC PROCOLIPASE AS DETERMINED FROM 1H HOMONUCLEAR TWO-AND THREE-DIMENSIONAL NMR | | Descriptor: | PORCINE PANCREATIC PROCOLIPASE B | | Authors: | Breg, J.N, Sarda, L, Cozzone, P.J, Rugani, N, Boelens, R, Kaptein, R. | | Deposit date: | 1994-06-08 | | Release date: | 1994-12-20 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic procolipase as determined from 1H homonuclear two-dimensional and three-dimensional NMR.
Eur.J.Biochem., 227, 1995
|
|
1PG5
 
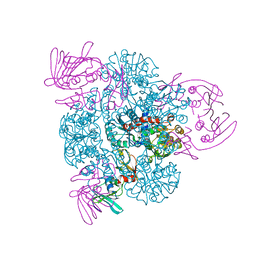 | | CRYSTAL STRUCTURE OF THE UNLIGATED (T-STATE) ASPARTATE TRANSCARBAMOYLASE FROM THE EXTREMELY THERMOPHILIC ARCHAEON SULFOLOBUS ACIDOCALDARIUS | | Descriptor: | Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | Authors: | De Vos, D, Van Petegem, F, Remaut, H, Legrain, C, Glansdorff, N, Van Beeumen, J.J. | | Deposit date: | 2003-05-27 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of T State Aspartate Carbamoyltransferase of the Hyperthermophilic Archaeon Sulfolobus acidocaldarius.
J.Mol.Biol., 339, 2004
|
|
1T1W
 
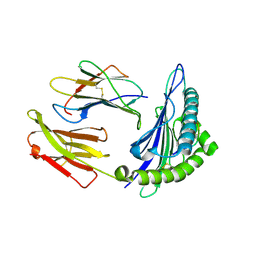 | | Structural basis for degenerate recognition of HIV peptide variants by cytotoxic lymphocyte, variant SL9-3F6I8V | | Descriptor: | Beta-2-microglobulin, GAG PEPTIDE, HLA class I histocompatibility antigen, ... | | Authors: | Martinez-Hackert, E, Anikeeva, N, Kalams, S.A, Walker, B.D, Hendrickson, W.A, Sykulev, Y. | | Deposit date: | 2004-04-19 | | Release date: | 2005-09-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Degenerate Recognition of Natural HIV Peptide Variants by Cytotoxic Lymphocytes.
J.Biol.Chem., 281, 2006
|
|
1SYV
 
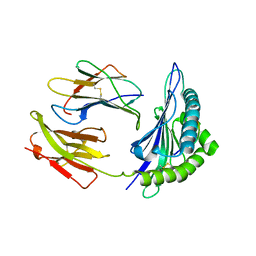 | | HLA-B*4405 complexed to the dominant self ligand EEFGRAYGF | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, major histocompatibility complex, ... | | Authors: | Zernich, D, Purcell, A.W, Macdonald, W.A, Kjer-Nielsen, L, Ely, L.K, Laham, N, Crockford, T, Mifsud, N.A, Tait, B.D, Holdsworth, R, Brooks, A.G, Bottomley, S.P, Beddoe, T, Peh, C.A, Rossjohn, J, McCluskey, J. | | Deposit date: | 2004-04-02 | | Release date: | 2004-10-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion
J.Exp.Med., 200, 2004
|
|
1T20
 
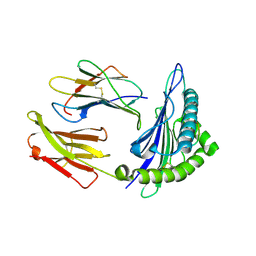 | | Structural basis for degenerate recognition of HIV peptide variants by cytotoxic lymphocyte, variant SL9-6I | | Descriptor: | Beta-2-microglobulin, GAG PEPTIDE, HLA class I histocompatibility antigen, ... | | Authors: | Martinez-Hackert, E, Anikeeva, N, Kalams, S.A, Walker, B.D, Hendrickson, W.A, Sykulev, Y. | | Deposit date: | 2004-04-19 | | Release date: | 2005-09-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Degenerate Recognition of Natural HIV Peptide Variants by Cytotoxic Lymphocytes.
J.Biol.Chem., 281, 2006
|
|
1SSE
 
 | |
1T4I
 
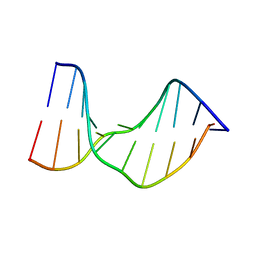 | | Crystal Structure of a DNA Decamer Containing a Thymine-dimer | | Descriptor: | 5'-D(*CP*GP*AP*AP*TP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*TP*TP*AP*AP*TP*TP*CP*G)-3' | | Authors: | Park, H, Zhang, K, Ren, Y, Nadji, S, Sinha, N, Taylor, J.S, Kang, C. | | Deposit date: | 2004-04-29 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a DNA Decamer Containing a cis-syn Thymine-dimer
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3VHV
 
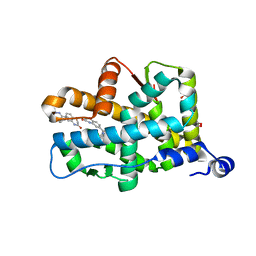 | | Mineralocorticoid receptor ligand-binding domain with non-steroidal antagonist | | Descriptor: | 1,2-ETHANEDIOL, 6-[(1E)-2-phenyl-N-(3-sulfanyl-4H-1,2,4-triazol-4-yl)ethanimidoyl]-2H-1,4-benzoxazin-3(4H)-one, 6-[(7S)-7-phenyl-7H-[1,2,4]triazolo[3,4-b][1,3,4]thiadiazin-6-yl]-2H-1,4-benzoxazin-3(4H)-one, ... | | Authors: | Sogabe, S, Habuka, N. | | Deposit date: | 2011-09-07 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identification of Benzoxazin-3-one Derivatives as Novel, Potent, and Selective Nonsteroidal Mineralocorticoid Receptor Antagonists
J.Med.Chem., 54, 2011
|
|
1T7H
 
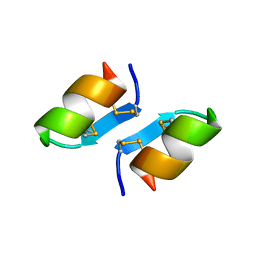 | | X-ray structure of [Lys(-2)-Arg(-1)-des(17-21)]-endothelin-1 peptide | | Descriptor: | Endothelin-1 | | Authors: | Hoh, F, Cerdan, R, Kaas, Q, Nishi, Y, Chiche, L, Kubo, S, Chino, N, Kobayashi, Y, Dumas, C, Aumelas, A. | | Deposit date: | 2004-05-10 | | Release date: | 2004-12-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | High-resolution X-ray structure of the unexpectedly stable dimer of the [Lys(-2)-Arg(-1)-des(17-21)]endothelin-1 peptide
Biochemistry, 43, 2004
|
|
