7QP6
 
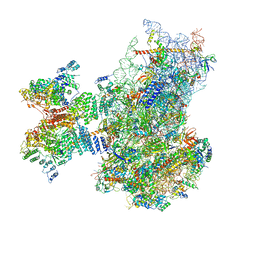 | | Structure of the human 48S initiation complex in open state (h48S AUG open) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Yi, S.-H, Petrychenko, V, Schliep, J.E, Goyal, A, Linden, A, Chari, A, Urlaub, H, Stark, H, Rodnina, M.V, Adio, S, Fischer, N. | | Deposit date: | 2022-01-03 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Conformational rearrangements upon start codon recognition in human 48S translation initiation complex.
Nucleic Acids Res., 50, 2022
|
|
7QP7
 
 | | Structure of the human 48S initiation complex in closed state (h48S AUG closed) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Yi, S.-H, Petrychenko, V, Schliep, J.E, Goyal, A, Linden, A, Chari, A, Urlaub, H, Stark, H, Rodnina, M.V, Adio, S, Fischer, N. | | Deposit date: | 2022-01-03 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Conformational rearrangements upon start codon recognition in human 48S translation initiation complex.
Nucleic Acids Res., 50, 2022
|
|
1UCR
 
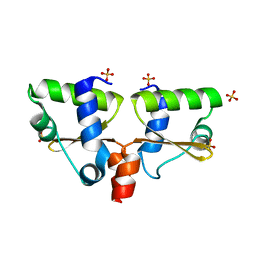 | | Three-dimensional crystal structure of dissimilatory sulfite reductase D (DsrD) | | Descriptor: | Protein dsvD, SULFATE ION | | Authors: | Mizuno, N, Voordouw, G, Miki, K, Sarai, A, Higuchi, Y. | | Deposit date: | 2003-04-18 | | Release date: | 2003-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of Dissimilatory Sulfite Reductase D (DsrD) Protein-Possible Interaction with B- and Z-DNA by Its Winged-Helix Motif
STRUCTURE, 11, 2003
|
|
7QDG
 
 | | SARS-CoV-2 S protein S:A222V + S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
7QDH
 
 | | SARS-CoV-2 S protein S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
6VPV
 
 | | Trimeric Photosystem I from the High-Light Tolerant Cyanobacteria Cyanobacterium Aponinum | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Dobson, Z, Toporik, H, Vaughn, N, Lin, S, Williams, D, Fromme, P, Mazor, Y. | | Deposit date: | 2020-02-04 | | Release date: | 2021-08-04 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The structure of photosystem I from a high-light tolerant Cyanobacteria.
Elife, 10, 2021
|
|
6VTP
 
 | |
6VTO
 
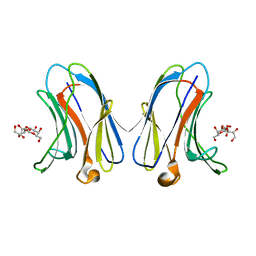 | | Crystal structure of human Galectin-7 in complex with 4-O-beta-D-Galactopyranosyl-D-glucose | | Descriptor: | (2~{R},3~{R},4~{R},5~{R})-4-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-2,3,5, 6-tetrakis(oxidanyl)hexanal, Galectin-7 | | Authors: | Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2020-02-13 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Perturbing dimer interactions and allosteric communication modulates the immunosuppressive activity of human galectin-7.
J.Biol.Chem., 297, 2021
|
|
6VTS
 
 | |
6VTQ
 
 | |
6VTR
 
 | | Crystal structure of G16S human Galectin-7 mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Galectin-7 | | Authors: | Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2020-02-13 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Perturbing dimer interactions and allosteric communication modulates the immunosuppressive activity of human galectin-7.
J.Biol.Chem., 297, 2021
|
|
6WCA
 
 | | Human closed state TMEM175 in KCl | | Descriptor: | Endosomal/lysosomal potassium channel TMEM175, POTASSIUM ION | | Authors: | Oh, S, Paknejad, N, Hite, R.K. | | Deposit date: | 2020-03-30 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Gating and selectivity mechanisms for the lysosomal K + channel TMEM175.
Elife, 9, 2020
|
|
7PQQ
 
 | |
7PQG
 
 | |
6WC9
 
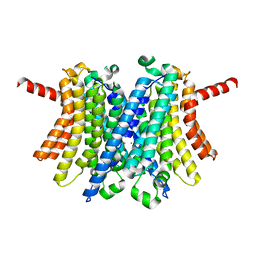 | | Human open state TMEM175 in KCl | | Descriptor: | Endosomal/lysosomal potassium channel TMEM175, POTASSIUM ION | | Authors: | Oh, S, Paknejad, N, Hite, R.K. | | Deposit date: | 2020-03-30 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Gating and selectivity mechanisms for the lysosomal K + channel TMEM175.
Elife, 9, 2020
|
|
6WHF
 
 | | class C beta-lactamase from Escherichia coli in complex with cephalothin | | Descriptor: | (2R)-5-[(acetyloxy)methyl]-2-{(1R)-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | class C beta-lactamase from Escherichia coli in complex with Cephalothin
To Be Published
|
|
6WHS
 
 | | GluN1b-GluN2B NMDA receptor in non-active 1 conformation at 3.95 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6WHV
 
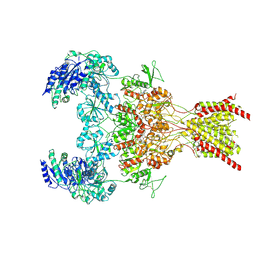 | | GluN1b-GluN2B NMDA receptor in complex with SDZ 220-040 and L689,560, class 2 | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, (2S)-2-amino-3-[2',4'-dichloro-4-hydroxy-5-(phosphonomethyl)biphenyl-3-yl]propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6WIF
 
 | | Class C beta-lactamase from Acinetobacter baumannii in complex with 4-(Ethyl(methyl)carbamoyl)phenyl boronic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase, ... | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | class C beta-lactamase from Acinetobacter baumannii in complex with 4-(Ethyl(methyl)carbamoyl)phenyl boronic acid
To Be Published
|
|
6WJ7
 
 | | The structure of NTMT1 in complex with compound C2A | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](ethyl)amino}-5'-deoxyadenosine, GLY-PRO-LYS-ARG-ILE-ALA-NH2, N-terminal Xaa-Pro-Lys N-methyltransferase 1 | | Authors: | Srinivasan, K, Chen, D, Huang, R, Noinaj, N. | | Deposit date: | 2020-04-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Probing the Plasticity in the Active Site of Protein N-terminal Methyltransferase 1 Using Bisubstrate Analogues.
J.Med.Chem., 63, 2020
|
|
7PB5
 
 | | Crystal structure of CD73 in complex with UMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-30 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7PBB
 
 | |
7PBA
 
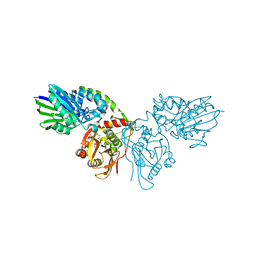 | | Crystal structure of CD73 in complex with IMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-08-01 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7PD9
 
 | |
7PCP
 
 | |
