7NEI
 
 | |
2KZB
 
 | | Solution structure of alpha-mannosidase binding domain of Atg19 | | Descriptor: | Autophagy-related protein 19 | | Authors: | Watanabe, Y, Noda, N, Kumeta, H, Suzuki, K, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2010-06-15 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Selective transport of alpha-mannosidase by autophagic pathways: structural basis for cargo recognition by Atg19 and Atg34.
J.Biol.Chem., 285, 2010
|
|
3VHV
 
 | | Mineralocorticoid receptor ligand-binding domain with non-steroidal antagonist | | Descriptor: | 1,2-ETHANEDIOL, 6-[(1E)-2-phenyl-N-(3-sulfanyl-4H-1,2,4-triazol-4-yl)ethanimidoyl]-2H-1,4-benzoxazin-3(4H)-one, 6-[(7S)-7-phenyl-7H-[1,2,4]triazolo[3,4-b][1,3,4]thiadiazin-6-yl]-2H-1,4-benzoxazin-3(4H)-one, ... | | Authors: | Sogabe, S, Habuka, N. | | Deposit date: | 2011-09-07 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identification of Benzoxazin-3-one Derivatives as Novel, Potent, and Selective Nonsteroidal Mineralocorticoid Receptor Antagonists
J.Med.Chem., 54, 2011
|
|
2L5Z
 
 | | NMR structure of the A730 loop of the Neurospora VS ribozyme | | Descriptor: | RNA (26-MER) | | Authors: | Desjardins, G, Bonneau, E, Girard, N, Boisbouvier, J, Legault, P. | | Deposit date: | 2010-11-10 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the A730 loop of the Neurospora VS ribozyme: insights into the formation of the active site.
Nucleic Acids Res., 39, 2011
|
|
7AA9
 
 | | Structure of SCOC pT13/pT15 LIR motif bound to GABARAPL1 | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, pT13/PT15 SCOC LIR | | Authors: | Lee, R, Mouilleron, S, Wirth, M, Zhang, W, O Reilly, N, Dhira, J, Tooze, S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity.
J.Mol.Biol., 433, 2021
|
|
7AA8
 
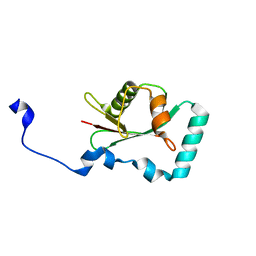 | | Structure of SCOC LIR bound to GABARAP | | Descriptor: | Chimera made of SCOC (6-23) + linker (GS) + GABARAP,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Lee, R, Mouilleron, S, Wirth, M, Zhang, W, O Reilly, N, Dhira, J, Tooze, S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity.
J.Mol.Biol., 433, 2021
|
|
7AA7
 
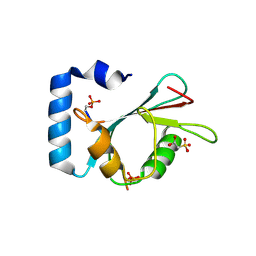 | | Structure of SCOC pS12/pS18 LIR motif bound to GABARAPL1 | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, pS12/pS18 SCOC LIR, sulfoacetic acid | | Authors: | Lee, R, Mouilleron, S, Wirth, M, Zhang, W, O Reilly, N, Dhira, J, Tooze, S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity.
J.Mol.Biol., 433, 2021
|
|
7Z3U
 
 | | Crystal structure of SARS-CoV-2 Main Protease after incubation with Sulfo-Calpeptin | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, Calpetin, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7Z58
 
 | | Crystal structure of human Cathepsin L in complex with covalently bound Calpeptin | | Descriptor: | 1,2-ETHANEDIOL, Calpeptin, Cathepsin L, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7Z3T
 
 | | Crystal structure of apo human Cathepsin L | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
2LAH
 
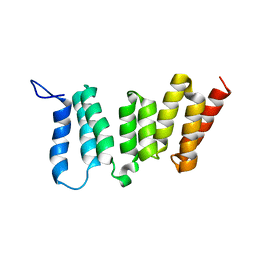 | | Solution NMR Structure of Mitotic checkpoint serine/threonine-protein kinase BUB1 N-terminal domain from Homo sapiens, Northeast Structural Genomics Consortium Target HR5460A (Methods Development) | | Descriptor: | Mitotic checkpoint serine/threonine-protein kinase BUB1 | | Authors: | Liu, G, Xiao, R, Lee, H, Hamilton, K, Acton, T.B, Ciccosanti, C, Everett, J.K, Shastry, R.T, Huang, Y.J, Montelione, G.T, Northeast Structural Genomics Consortium, n, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target HR5460A
To be Published
|
|
2LTB
 
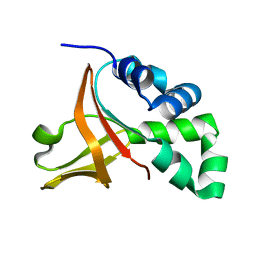 | | Wild-type FAS1-4 | | Descriptor: | Transforming growth factor-beta-induced protein ig-h3 | | Authors: | Underhaug, J, Nielsen, N, Runager, K. | | Deposit date: | 2012-05-16 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Mutation in transforming growth factor beta induced protein associated with granular corneal dystrophy type 1 reduces the proteolytic susceptibility through local structural stabilization.
Biochim.Biophys.Acta, 1834, 2013
|
|
2L45
 
 | | C-terminal zinc knuckle of the HIVNCp7 with DNA | | Descriptor: | C-TERMINAL ZINC KNUCLE OF THE HIV-NCP7, DNA (5'-D(P*TP*AP*CP*GP*CP*C)-3'), ZINC ION | | Authors: | Quintal, S, Viegas, A, Cabrita, E, Farrell, N, Erhardt, S. | | Deposit date: | 2010-10-01 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Platinated DNA affects zinc finger conformation. Interaction of a platinated single-stranded oligonucleotide and the C-terminal zinc finger of nucleocapsid protein HIVNCp7.
Biochemistry, 51, 2012
|
|
2L8I
 
 | | A biocompatible backbone modification? - Structure and dynamics of a triazole-linked DNA duplex | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G*(2L8)P*TP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*AP*AP*AP*CP*GP*TP*CP*G)-3') | | Authors: | El-Sagheer, A, Brown, T, Ernsting, N, Dehmel, L, Griesinger, C, Mugge, C. | | Deposit date: | 2011-01-13 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of Triazole-Linked DNA: Biocompatibility Explained.
Chemistry, 17, 2011
|
|
7NPW
 
 | |
2L3E
 
 | | Solution structure of P2a-J2a/b-P2b of human telomerase RNA | | Descriptor: | 35-MER | | Authors: | Zhang, Q, Kim, N, Peterson, R.D, Wang, Z, Feigon, J. | | Deposit date: | 2010-09-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Inaugural Article: Structurally conserved five nucleotide bulge determines the overall topology of the core domain of human telomerase RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2L4R
 
 | | NMR solution structure of the N-terminal PAS domain of hERG | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Gayen, N, Li, Q, Chen, A.S, Huang, Q, Raida, M, Kang, C. | | Deposit date: | 2010-10-13 | | Release date: | 2010-12-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the N-terminal domain of hERG and its interaction with the S4-S5 linker.
Biochem.Biophys.Res.Commun., 403, 2010
|
|
2LF4
 
 | |
7N0W
 
 | |
7N0Y
 
 | |
2L2L
 
 | |
2L46
 
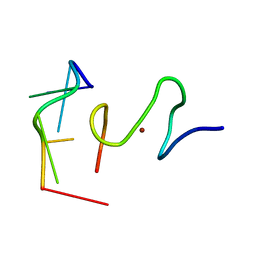 | | C-terminal zinc finger of the HIVNCp7 with platinated DNA | | Descriptor: | C-TERMINAL ZINC KNUCLE OF THE HIV-NCP7, DNA (5'-D(P*TP*AP*CP*GP*CP*C)-3'), ZINC ION | | Authors: | Quintal, S, Viegas, A, Cabrita, E, Farrell, N, Erhardt, S. | | Deposit date: | 2010-10-01 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Platinated DNA affects zinc finger conformation. Interaction of a platinated single-stranded oligonucleotide and the C-terminal zinc finger of nucleocapsid protein HIVNCp7.
Biochemistry, 51, 2012
|
|
7N43
 
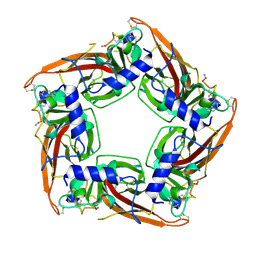 | |
7NBV
 
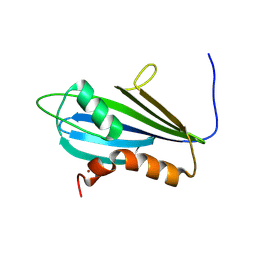 | | Structure of 2A protein from Theilers murine encephalomyelitis virus (TMEV) | | Descriptor: | BROMIDE ION, Capsid protein VP0 | | Authors: | Hill, C.H, Cook, G.M, Napthine, S, Kibe, A, Brown, K, Caliskan, N, Firth, A.E, Graham, S.C, Brierley, I. | | Deposit date: | 2021-01-28 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Investigating molecular mechanisms of 2A-stimulated ribosomal pausing and frameshifting in Theilovirus.
Nucleic Acids Res., 49, 2021
|
|
7NHF
 
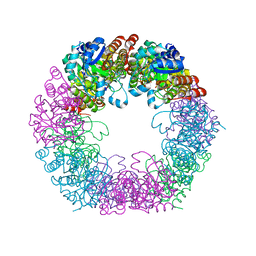 | | Crystal structure of Arabidopsis thaliana Pdx1K166R | | Descriptor: | PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3 | | Authors: | Rodrigues, M.J, Zhang, Y, Bolton, R, Evans, G, Giri, N, Royant, A, Begley, T, Ealick, S.E, Tews, I. | | Deposit date: | 2021-02-10 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Trapping and structural characterisation of a covalent intermediate in vitamin B 6 biosynthesis catalysed by the Pdx1 PLP synthase.
Rsc Chem Biol, 3, 2022
|
|
