6FWW
 
 | | GFP/KKK. A redesigned GFP with improved solubility | | Descriptor: | Green fluorescent protein | | Authors: | Varejao, N, Lascorz, J, Gil-Garcia, M, Diaz-Caballero, M, Navarro, S, Ventura, S, Reverter, D. | | Deposit date: | 2018-03-07 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.131 Å) | | Cite: | Combining Structural Aggregation Propensity and Stability Predictions To Redesign Protein Solubility.
Mol. Pharm., 15, 2018
|
|
6FUH
 
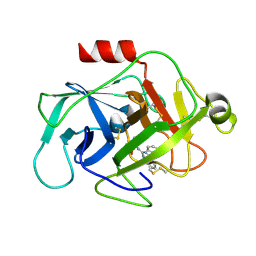 | | Complement factor D in complex with the inhibitor (4-((3-(aminomethyl)phenyl)amino)quinazolin-2-yl)-L-valine | | Descriptor: | (2~{S})-2-[[4-[[3-(aminomethyl)phenyl]amino]quinazolin-2-yl]amino]-3-methyl-butanoic acid, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
6CS1
 
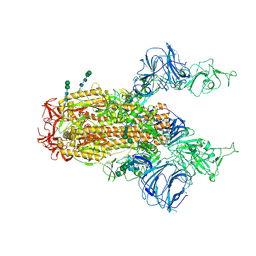 | | SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, two S1 CTDs in an upwards conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
1AHK
 
 | | DER F 2, THE MAJOR MITE ALLERGEN FROM DERMATOPHAGOIDES FARINAE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DER F 2 | | Authors: | Ichikawa, S, Hatanaka, H, Yuuki, T, Iwamoto, N, Ogura, K, Okumura, Y, Inagaki, F. | | Deposit date: | 1997-04-07 | | Release date: | 1998-04-08 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Der f 2, the major mite allergen for atopic diseases.
J.Biol.Chem., 273, 1998
|
|
5DE5
 
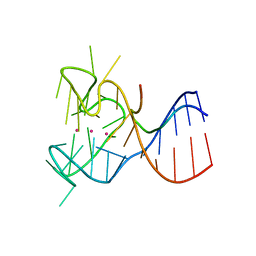 | | Crystal structure of the complex between human FMRP RGG motif and G-quadruplex RNA. | | Descriptor: | Fragile X mental retardation protein 1, POTASSIUM ION, sc1 | | Authors: | Vasilyev, N, Polonskaia, A, Darnell, J.C, Darnell, R.B, Patel, D.J, Serganov, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.0011 Å) | | Cite: | Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6CIH
 
 | | Crystal structure of a group II intron lariat in the post-catalytic state | | Descriptor: | IRIDIUM HEXAMMINE ION, MAGNESIUM ION, RNA (5'-R(P*UP*GP*UP*UP*UP*AP*UP*UP*AP*AP*AP*AP*AP*C*-3'), ... | | Authors: | Chan, R.T, Peters, J.K, Robart, A.R, Wiryaman, T, Rajashankar, K.R, Toor, N. | | Deposit date: | 2018-02-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.676 Å) | | Cite: | Structural basis for the second step of group II intron splicing.
Nat Commun, 9, 2018
|
|
6V70
 
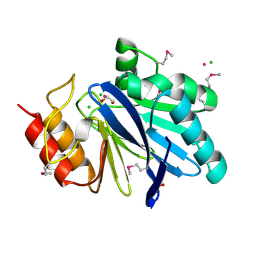 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Cadmium in the Active Site | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CADMIUM ION, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-06 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica.
To Be Published
|
|
6G1V
 
 | | Crystal structure of Torpedo Californica acetylcholinesterase in complex with 12-Amino-3-chloro-6,7,10,11-tetrahydro-5,9-dimethyl-7,11-methanocycloocta[b]quinolin-5-ium | | Descriptor: | 12-Amino-3-chloro-6,7,10,11-tetrahydro-5,9-dimethyl-7,11-methanocycloocta[b]quinolin-5-ium, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Coquelle, N, Colletier, J.P. | | Deposit date: | 2018-03-22 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Increasing Polarity in Tacrine and Huprine Derivatives: Potent Anticholinesterase Agents for the Treatment of Myasthenia Gravis.
Molecules, 23, 2018
|
|
5DA8
 
 | | Crystal structure of chaperonin GroEL from | | Descriptor: | 60 kDa chaperonin, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Chang, C, Marshall, N, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-08-19 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of chaperonin GroEL from
To Be Published
|
|
5DGJ
 
 | | 1.0A resolution structure of Norovirus 3CL protease in complex an oxadiazole-based, cell permeable macrocyclic (20-mer) inhibitor | | Descriptor: | 3C-LIKE PROTEASE, tert-butyl [(4S,7S,10S)-7-(cyclohexylmethyl)-10-(hydroxymethyl)-5,8,13-trioxo-22-oxa-6,9,14,20,21-pentaazabicyclo[17.2.1]docosa-1(21),19-dien-4-yl]carbamate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Alliston, K.R, Weerawarna, P.M, Kankanamalage, A.C.G, Lushington, G.H, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2015-08-27 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Oxadiazole-Based Cell Permeable Macrocyclic Transition State Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 59, 2016
|
|
6FRK
 
 | | Structure of a prehandover mammalian ribosomal SRP and SRP receptor targeting complex | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kobayashi, K, Jomaa, A, Ban, N. | | Deposit date: | 2018-02-16 | | Release date: | 2018-03-28 | | Last modified: | 2018-05-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a prehandover mammalian ribosomal SRP·SRP receptor targeting complex.
Science, 360, 2018
|
|
6PBC
 
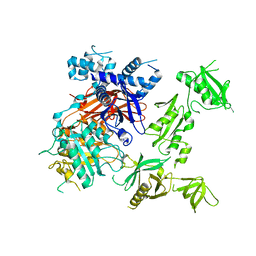 | | Structural basis for the activation of PLC-gamma isozymes by phosphorylation and cancer-associated mutations | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma,1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1, CALCIUM ION, SODIUM ION | | Authors: | Hajicek, N, Sondek, J. | | Deposit date: | 2019-06-13 | | Release date: | 2020-01-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis for the activation of PLC-gamma isozymes by phosphorylation and cancer-associated mutations.
Elife, 8, 2019
|
|
5CQ5
 
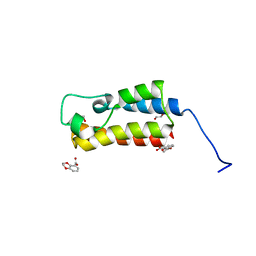 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 2,3-Ethylenedioxybenzoic Acid (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 2,3-Ethylenedioxybenzoic Acid (SGC - Diamond I04-1 fragment screening)
To be published
|
|
6G8G
 
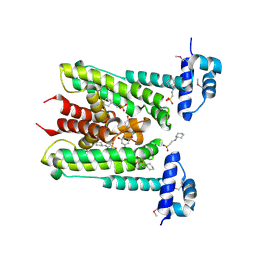 | | Flavonoid-responsive Regulator FrrA in complex with Genistein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GENISTEIN, TetR/AcrR family transcriptional regulator | | Authors: | Werner, N, Hoppen, J, Palm, G, Werten, S, Goettfert, M, Hinrichs, W. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The induction mechanism of the flavonoid-responsive regulator FrrA.
Febs J., 2021
|
|
6FZJ
 
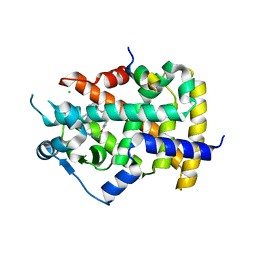 | | PPAR gamma mutant complex | | Descriptor: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, CHLORIDE ION, IODIDE ION, ... | | Authors: | Rochel, N. | | Deposit date: | 2018-03-14 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Recurrent activating mutations of PPAR gamma associated with luminal bladder tumors.
Nat Commun, 10, 2019
|
|
6P6D
 
 | | HUMAN IGG1 FC FRAGMENT, C239 INSERTION MUTANT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CYSTEINE, IgG1 Fc fragment | | Authors: | Gallagher, D.T, Dimasi, N, McCullough, C. | | Deposit date: | 2019-06-03 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure and Dynamics of a Site-Specific Labeled Fc Fragment with Altered Effector Functions.
Pharmaceutics, 11, 2019
|
|
1PS3
 
 | | Golgi alpha-mannosidase II in complex with kifunensine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-mannosidase II, ... | | Authors: | Shah, N, Kuntz, D.A, Rose, D.R. | | Deposit date: | 2003-06-20 | | Release date: | 2003-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of Kifunensine and 1-Deoxymannojirimycin Binding to Class I and II alpha-Mannosidases Demonstrates Different Saccharide Distortions in Inverting and Retaining Catalytic Mechanisms
Biochemistry, 42, 2003
|
|
6G5P
 
 | | Crystal structure of human SP100 in complex with bromodomain-focused fragment FM009493b 2,3-Dimethoxy-2,3-dimethyl-2,3-dihydro-1,4-benzodioxin-6-amine | | Descriptor: | (2~{R},3~{R})-2,3-dimethoxy-2,3-dimethyl-1,4-benzodioxin-6-amine, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Talon, R.P.H, Krojer, T, Tallant, C, Nunez-Alonso, G, Fairhead, M, Szykowska, A, Collins, P, Pearce, N.M, Ng, J, MacLean, E, Wright, N, Douangamath, A, Brandao-Neto, J, Burgess-Brown, N, Huber, K, Knapp, S, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2018-03-29 | | Release date: | 2018-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identifying small molecule binding sites for epigenetic proteins at domain-domain interfaces
Biorxiv, 2018
|
|
1PVV
 
 | |
6CMC
 
 | |
6PDN
 
 | | Human PIM1 bound to benzothiophene inhibitor 292 | | Descriptor: | 4-{5-[(3-aminopropyl)carbamoyl]thiophen-2-yl}-1-benzothiophene-2-carboxylic acid, GLYCEROL, Serine/threonine-protein kinase pim-1, ... | | Authors: | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human PIM1
To Be Published
|
|
6PDO
 
 | | Human PIM1 bound to benzothiophene inhibitor 354 | | Descriptor: | 4-{5-[(3-aminopropyl)carbamoyl]thiophen-2-yl}-1-benzothiophene-2-carboxamide, Peptide, Serine/threonine-protein kinase pim-1 | | Authors: | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human PIM1
To Be Published
|
|
6PPR
 
 | | Cryo-EM structure of AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP and DNA | | Descriptor: | ATP-dependent DNA helicase (UvrD/REP), DNA (70-MER), IRON/SULFUR CLUSTER, ... | | Authors: | Jia, N, Unciuleac, M, Shuman, S, Patel, D.J. | | Deposit date: | 2019-07-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6UX3
 
 | | Crystal structure of acetoin dehydrogenase from Enterobacter cloacae | | Descriptor: | Acetoin dehydrogenase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Chang, C, Skarina, T, Mesa, N, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-11-06 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Crystal structure of acetoin dehydrogenase from Enterobacter cloacae
To Be Published
|
|
6V0C
 
 | | Lipophilic Envelope-spanning Tunnel B (LetB), Model 1 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-11-18 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
