1N3B
 
 | | Crystal Structure of Dephosphocoenzyme A kinase from Escherichia coli | | Descriptor: | Dephospho-CoA kinase, SULFATE ION | | Authors: | O'Toole, N, Barbosa, J.A.R.G, Li, Y, Hung, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-10-25 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Trimeric Form of Dephosphocoenzyme A Kinase from Escherichia coli
Protein Sci., 12, 2003
|
|
4ZU2
 
 | | Pseudomonas aeruginosa AtuE | | Descriptor: | IODIDE ION, Putative isohexenylglutaconyl-CoA hydratase | | Authors: | Poudel, N, Pfannstiel, J, Simon, O, Walter, N, Jendrossek, D, Papageorgiou, A.C. | | Deposit date: | 2015-05-15 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Pseudomonas aeruginosa Isohexenyl Glutaconyl Coenzyme A Hydratase (AtuE) Is Upregulated in Citronellate-Grown Cells and Belongs to the Crotonase Family.
Appl.Environ.Microbiol., 81, 2015
|
|
1JGN
 
 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip2 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein 2 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-26 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
2RVO
 
 | | Solution structure of a reverse transcriptase recognition site of a LINE RNA from zebrafish | | Descriptor: | RNA (34-MER) | | Authors: | Otsu, M, Norose, N, Arai, N, Terao, R, Kajikawa, M, Okada, N, Kawai, G. | | Deposit date: | 2016-02-03 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a reverse transcriptase recognition site of a LINE RNA from zebrafish.
J. Biochem., 162, 2017
|
|
4WZN
 
 | | CRYSTAL STRUCTURE OF THE 2B PROTEIN SOLUBLE DOMAIN FROM HEPATITIS A VIRUS | | Descriptor: | GLYCEROL, Genome polyprotein | | Authors: | Garriga, D, Vives-Adrian, L, Buxaderas, M, Ferreira-da-Silva, F, Almeida, B, Macedo-Ribeiro, S, Pereira, P.J, Verdaguer, N. | | Deposit date: | 2014-11-20 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Host Membrane Remodeling Induced by Protein 2B of Hepatitis A Virus.
J.Virol., 89, 2015
|
|
5V4E
 
 | | Engineered human IgG Fc domain glyco801 (Fc801) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, W, Marshall, N, Zhang, Y.J. | | Deposit date: | 2017-03-09 | | Release date: | 2017-06-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.216 Å) | | Cite: | IgG Fc domains that bind C1q but not effector Fc gamma receptors delineate the importance of complement-mediated effector functions.
Nat. Immunol., 18, 2017
|
|
1NBB
 
 | | N-BUTYLISOCYANIDE BOUND RHODOBACTER CAPSULATUS CYTOCHROME C' | | Descriptor: | CYTOCHROME C', N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tahirov, T.H, Misaki, S, Meyer, T.E, Cusanovich, M.A, Higuchi, Y, Yasuoka, N. | | Deposit date: | 1996-03-18 | | Release date: | 1996-08-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Concerted movement of side chains in the haem vicinity observed on ligand binding in cytochrome c' from rhodobacter capsulatus.
Nat.Struct.Biol., 3, 1996
|
|
4Z5T
 
 | | The nucleosome containing human H3.5 | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Urahama, T, Harada, A, Maehara, K, Horikoshi, N, Sato, K, Sato, Y, Shiraishi, K, Sugino, N, Osakabe, A, Tachiwana, H, Kagawa, W, Kimura, H, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2015-04-03 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Histone H3.5 forms an unstable nucleosome and accumulates around transcription start sites in human testis.
Epigenetics Chromatin, 9, 2016
|
|
4TNH
 
 | | RT XFEL structure of Photosystem II in the dark state at 4.9 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.900007 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
2RPA
 
 | | The solution structure of N-terminal domain of microtubule severing enzyme | | Descriptor: | Katanin p60 ATPase-containing subunit A1 | | Authors: | Iwaya, N, Kuwahara, Y, Unzai, S, Nagata, T, Tomii, K, Goda, N, Tochio, H, Shirakawa, M, Hiroaki, H. | | Deposit date: | 2008-05-13 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A common substrate recognition mode conserved between katanin P60 and VPS4 governs microtubule severing and membrane skeleton reorganization
J.Biol.Chem., 285, 2010
|
|
2ISM
 
 | | Crystal structure of the putative oxidoreductase (glucose dehydrogenase) (TTHA0570) from thermus theromophilus HB8 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative oxidoreductase | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Ebihara, A, Shinkai, A, Nakagawa, N, Shimizu, N, Yamamoto, M, Kuramitsu, S, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-18 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Putative Oxidoreductase (Glucose Dehydrogenase) (TTHA0570) from Thermus Theromophilus HB8
To be Published
|
|
2RRE
 
 | | Structure and function of the N-terminal nucleolin binding domain of nuclear valocine containing protein like 2 (NVL2) harboring a nucleolar localization signal | | Descriptor: | Putative uncharacterized protein | | Authors: | Fujiwara, Y, Fujiwara, K, Goda, N, Iwaya, N, Tenno, T, Shirakawa, M, Hiroaki, H. | | Deposit date: | 2010-08-03 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the N-terminal nucleolin binding domain of nuclear valosin-containing protein-like 2 (NVL2) harboring a nucleolar localization signal
J.Biol.Chem., 286, 2011
|
|
4TNL
 
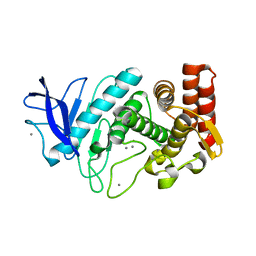 | | 1.8 A resolution room temperature structure of Thermolysin recorded using an XFEL | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
1FFK
 
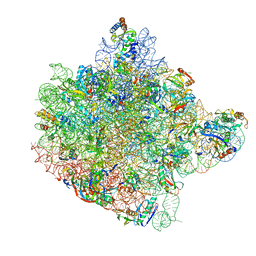 | | CRYSTAL STRUCTURE OF THE LARGE RIBOSOMAL SUBUNIT FROM HALOARCULA MARISMORTUI AT 2.4 ANGSTROM RESOLUTION | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Ban, N, Nissen, P, Hansen, J, Moore, P.B, Steitz, T.A. | | Deposit date: | 2000-07-25 | | Release date: | 2000-08-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The complete atomic structure of the large ribosomal subunit at 2.4 A resolution.
Science, 289, 2000
|
|
2IE1
 
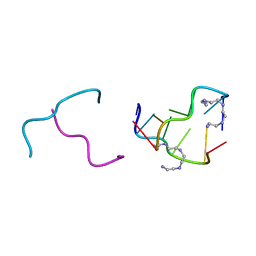 | | Polyamines stabilize left-handed Z-DNA. We found new type of polyamine which stabilize left-handed Z-DNA by X-ray crystallography | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DG)-3'), N-(2-AMINOETHYL)-N'-{2-[(2-AMINOETHYL)AMINO]ETHYL}ETHANE-1,2-DIAMINE | | Authors: | Ohishi, H, Odoko, M, Tsukamoto, K, Hiyama, Y, Maezaki, N, Grzeskowiak, K, Ishida, T, Tanaka, T, Okabe, N, Fukuyama, K. | | Deposit date: | 2006-09-16 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Polyamines stabilize left-handed Z-DNA. We found new type of polyamine which stabilize left-handed Z-DNA by X-ray crystallography
To be Published
|
|
1OSJ
 
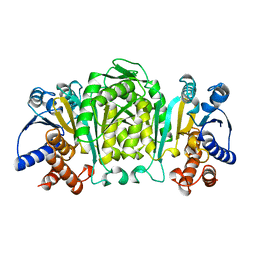 | | STRUCTURE OF 3-ISOPROPYLMALATE DEHYDROGENASE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Moriyama, H, Tanaka, N, Oshima, T. | | Deposit date: | 1996-10-22 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A mutation at the interface between domains causes rearrangement of domains in 3-isopropylmalate dehydrogenase.
Protein Eng., 10, 1997
|
|
1JH4
 
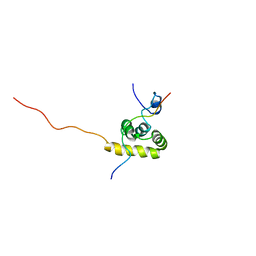 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip1 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein-1 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-27 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
1OI8
 
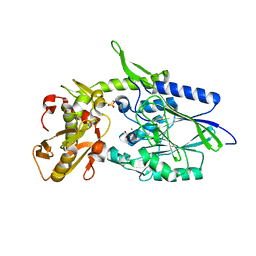 | | 5'-Nucleotidase (E. coli) with an Engineered Disulfide Bridge (P90C, L424C) | | Descriptor: | CARBONATE ION, MANGANESE (II) ION, PROTEIN USHA, ... | | Authors: | Schultz-Heienbrok, R, Maier, T, Straeter, N. | | Deposit date: | 2003-06-10 | | Release date: | 2004-06-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trapping a 96 Degree Domain Rotation in Two Distinct Conformations by Engineered Disulfide Bridges
Protein Sci., 13, 2004
|
|
1OIE
 
 | |
5DI0
 
 | | Crystal structure of Dln1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jia, N, Jiang, Y.L, Cheng, W, Wang, H.W, Zhou, C.Z, Chen, Y. | | Deposit date: | 2015-08-31 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for receptor recognition and pore formation of a zebrafish aerolysin-like protein.
Embo Rep., 17, 2016
|
|
1FFZ
 
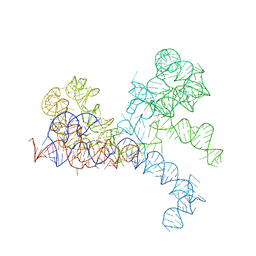 | | LARGE RIBOSOMAL SUBUNIT COMPLEXED WITH R(CC)-DA-PUROMYCIN | | Descriptor: | 23S RIBOSOMAL RNA, R(P*CP*C*)-D(P*A)-R(P*(PU)) | | Authors: | Nissen, P, Hansen, J, Ban, N, Moore, P.B, Steitz, T.A. | | Deposit date: | 2000-07-26 | | Release date: | 2000-08-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis of ribosome activity in peptide bond synthesis.
Science, 289, 2000
|
|
1FHO
 
 | | Solution Structure of the PH Domain from the C. Elegans Muscle Protein UNC-89 | | Descriptor: | UNC-89 | | Authors: | Blomberg, N, Baraldi, E, Sattler, M, Saraste, M, Nilges, M. | | Deposit date: | 2000-08-02 | | Release date: | 2000-10-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a PH domain from the C. elegans muscle protein UNC-89 suggests a novel function.
Structure Fold.Des., 8, 2000
|
|
1FPZ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF KINASE ASSOCIATED PHOSPHATASE (KAP) WITH A SUBSTITUTION OF THE CATALYTIC SITE CYSTEINE (CYS140) TO A SERINE | | Descriptor: | CYCLIN-DEPENDENT KINASE INHIBITOR 3, SULFATE ION | | Authors: | Song, H, Hanlon, N, Brown, N.R, Noble, M.E.M, Johnson, L.N, Barford, D. | | Deposit date: | 2000-09-01 | | Release date: | 2001-05-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphoprotein-protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2.
Mol.Cell, 7, 2001
|
|
4YM6
 
 | | Crystal structure of the human nucleosome containing 6-4PP (outside) | | Descriptor: | 145-MER DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Osakabe, A, Tachiwana, H, Kagawa, W, Horikoshi, N, Matsumoto, S, Hasegawa, M, Matsumoto, N, Toga, T, Yamamoto, J, Hanaoka, F, Thoma, N.H, Sugasawa, K, Iwai, S, Kurumizaka, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.514 Å) | | Cite: | Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome
Sci Rep, 5, 2015
|
|
5DYH
 
 | | Ti(IV) bound human serum transferrin | | Descriptor: | CARBONATE ION, CITRIC ACID, Serotransferrin, ... | | Authors: | Saxena, M, Sharma, S, Noinaj, N, Parks, T.B, Tinoco, A.D. | | Deposit date: | 2015-09-24 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.682 Å) | | Cite: | Unusual Synergism of Transferrin and Citrate in the Regulation of Ti(IV) Speciation, Transport, and Toxicity.
J.Am.Chem.Soc., 138, 2016
|
|
