1VCI
 
 | | Crystal structure of the ATP-binding cassette of multisugar transporter from Pyrococcus horikoshii OT3 complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, sugar-binding transport ATP-binding protein | | Authors: | Ose, T, Fujie, T, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2004-03-08 | | Release date: | 2004-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the ATP-binding cassette of multisugar transporter from Pyrococcus horikoshii OT3
Proteins, 57, 2004
|
|
1VDG
 
 | | Crystal structure of LIR1.01, one of the alleles of LIR1 | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily B member 1 | | Authors: | Shiroishi, M, Rasubala, L, Kuroki, K, Amano, K, Tsuchiya, N, Tokunaga, K, Kohda, D, Maenaka, K. | | Deposit date: | 2004-03-22 | | Release date: | 2005-08-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of LIR1.03, one of the alleles of LIR1
To be Published
|
|
1VE3
 
 | | Crystal structure of PH0226 protein from Pyrococcus horikoshii OT3 | | Descriptor: | S-ADENOSYLMETHIONINE, hypothetical protein PH0226 | | Authors: | Lokanath, N.K, Yamamoto, H, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-26 | | Release date: | 2005-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase from Pyrococcus horikoshii.
Acta Crystallogr.,Sect.F, 73, 2017
|
|
1VDZ
 
 | | Crystal structure of A-type ATPase catalytic subunit A from Pyrococcus horikoshii OT3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, A-type ATPase subunit A | | Authors: | Maegawa, Y, Morita, H, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2004-03-26 | | Release date: | 2005-06-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of A-type ATPase catalytic subunit A from Pyrococcus horikoshii OT3
To be Published
|
|
1VFI
 
 | | Solution Structure of Vanabin2 (RUH-017), a Vanadium-binding Protein from Ascidia sydneiensis samea | | Descriptor: | vanadium-binding protein 2 | | Authors: | Hamada, T, Hirota, H, Asanuma, M, Hayashi, F, Kobayashi, N, Ueki, T, Michibata, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-04-13 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Vanabin2, a Vanadium(IV)-Binding Protein from the Vanadium-Rich Ascidian Ascidia sydneiensis samea
J.Am.Chem.Soc., 127, 2005
|
|
2L5J
 
 | | structure of the spliceosomal phosphopeptide P140 (phosphorylated form) | | Descriptor: | U1 small nuclear ribonucleoprotein 70 kDa | | Authors: | Quinternet, M, Page, N, Schall, N, Strub, J, Chaloin, O, Decossas, M, Cung, M, van Dorsselaer, A, Briand, J, Muller, S. | | Deposit date: | 2010-11-02 | | Release date: | 2010-12-01 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The spliceosomal phosphopeptide P140 controls the lupus disease by interacting with the HSC70 protein and via a mechanism mediated by gammadelta T cells.
Plos One, 4, 2009
|
|
1W4F
 
 | | Peripheral-subunit from mesophilic, thermophilic and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1VGO
 
 | | Crystal Structure of Archaerhodopsin-2 | | Descriptor: | Archaerhodopsin 2, RETINAL, SULFATE ION, ... | | Authors: | Yoshimura, K, Enami, N, Murakami, M, Okumura, H, Ihara, K, Kouyama, T. | | Deposit date: | 2004-04-28 | | Release date: | 2005-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of archaerhodopsin-1 and -2: Common structural motif in archaeal light-driven proton pumps
J.Mol.Biol., 358, 2006
|
|
1VCK
 
 | | Crystal structure of ferredoxin component of carbazole 1,9a-dioxygenase of Pseudomonas resinovorans strain CA10 | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, HYDROSULFURIC ACID, ... | | Authors: | Nam, J.-W, Noguchi, H, Fujiomoto, Z, Mizuno, H, Fushinobu, S, Kobashi, N, Iwata, K, Yoshida, T, Habe, H, Yamane, H, Omori, T, Nojiri, H. | | Deposit date: | 2004-03-09 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the ferredoxin component of carbazole 1,9a-dioxygenase of Pseudomonas resinovorans strain CA10, a novel Rieske non-heme iron oxygenase system
PROTEINS, 58, 2005
|
|
1VDK
 
 | |
1WBF
 
 | | WINGED BEAN LECTIN, SACCHARIDE FREE FORM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Manoj, N, Srinivas, V.R, Suguna, K. | | Deposit date: | 1998-12-16 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of basic winged-bean lectin and a comparison with its saccharide-bound form.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1W78
 
 | | E.coli FolC in complex with DHPP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FOLC BIFUNCTIONAL PROTEIN, MAGNESIUM ION, ... | | Authors: | Mathieu, M, Debousker, G, Vincent, S, Viviani, F, Bamas-Jacques, N, Mikol, V. | | Deposit date: | 2004-08-31 | | Release date: | 2005-02-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Escherichia Coli Folc Structure Reveals an Unexpected Dihydrofolate Binding Site Providing an Attractive Target for Anti-Microbial Therapy
J.Biol.Chem., 280, 2005
|
|
1WB4
 
 | | S954A mutant of the feruloyl esterase module from clostridium thermocellum complexed with sinapinate | | Descriptor: | ACETIC ACID, CADMIUM ION, ENDO-1,4-BETA-XYLANASE Y, ... | | Authors: | Tarbouriech, N, Prates, J.A, Fontes, C, Davies, G.J. | | Deposit date: | 2004-10-30 | | Release date: | 2005-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Determinants of Substrate Specificity in the Feruloyl Esterase Module of Xylanase 10B from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1W18
 
 | | Crystal Structure of levansucrase from Gluconacetobacter diazotrophicus | | Descriptor: | LEVANSUCRASE, SULFATE ION | | Authors: | Martinez-Fleites, C, Ortiz-Lombardia, M, Pons, T, Tarbouriech, N, Taylor, E.J, Hernandez, L, Davies, G.J. | | Deposit date: | 2004-06-16 | | Release date: | 2005-05-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Levansucrase from the Gram- Negative Bacterium Gluconacetobacter Diazotrophicus.
Biochem.J., 390, 2005
|
|
7NFV
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Gunther, S, Reinke, P, Werner, N, Falke, S, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Sprenger, J, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Koua, F, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Ewert, W, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Ehrt, C, Rarey, M, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Hinrichs, W, Meents, A, Betzel, C. | | Deposit date: | 2021-02-07 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
2L5I
 
 | | structure of the spliceosomal phosphopeptide P140 (non-phosphorylated form) | | Descriptor: | U1 small nuclear ribonucleoprotein 70 kDa | | Authors: | Quinternet, M, Page, N, Schall, N, Strub, J, Chaloin, O, Decossas, M, Cung, M, van Dorsselaer, A, Briand, J, Muller, S. | | Deposit date: | 2010-11-02 | | Release date: | 2010-12-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The spliceosomal phosphopeptide P140 controls the lupus disease by interacting with the HSC70 protein and via a mechanism mediated by gammadelta T cells.
Plos One, 4, 2009
|
|
5HO3
 
 | | MamB | | Descriptor: | Magnetosome protein MamB | | Authors: | Zeytuni, N, Keren, N, Davidov, G, Zarivach, R. | | Deposit date: | 2016-01-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The dual role of MamB in magnetosome membrane assembly and magnetite biomineralization.
Mol. Microbiol., 107, 2018
|
|
6QSW
 
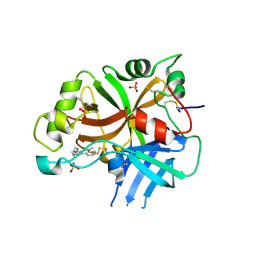 | | Complement factor B protease domain in complex with the reversible inhibitor N-(2-bromo-4-methylnaphthalen-1-yl)-4,5-dihydro-1H-imidazol-2-amine. | | Descriptor: | Complement factor B, SULFATE ION, ~{N}-(2-bromanyl-4-methyl-naphthalen-1-yl)-4,5-dihydro-1~{H}-imidazol-2-amine | | Authors: | Adams, C.M, Sellner, H, Ehara, T, Mac Sweeney, A, Crowley, M, Anderson, K, Karki, R, Mainolfi, N, Valeur, E, Sirockin, F, Gerhartz, B, Erbel, P, Hughes, N, Smith, T.M, Cumin, F, Argikar, U, Mogi, M, Sedrani, R, Wiesmann, C, Jaffee, B, Maibaum, J, Flohr, S, Harrison, R, Eder, J. | | Deposit date: | 2019-02-22 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Small-molecule factor B inhibitor for the treatment of complement-mediated diseases.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7CRZ
 
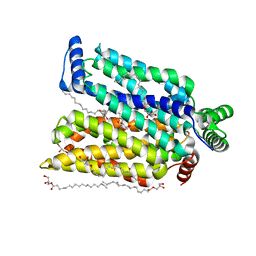 | | Crystal structure of human glucose transporter GLUT3 bound with C3361 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,3R,4S,5R,6R)-6-(hydroxymethyl)-4-undec-10-enoxy-oxane-2,3,5-triol, Solute carrier family 2, ... | | Authors: | Yuan, Y.Y, Zhang, S, Wang, N, Jiang, X, Yan, N. | | Deposit date: | 2020-08-14 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Orthosteric-allosteric dual inhibitors of PfHT1 as selective antimalarial agents.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6QSX
 
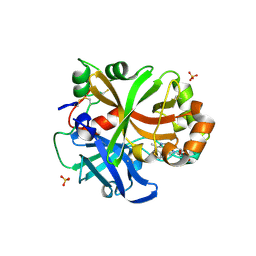 | | Complement factor B protease domain in complex with the reversible inhibitor ((2S,4S)-1-((5,7-dimethyl-1H-indol-4-yl)methyl)-4-methoxypiperidin-2-yl)methanol. | | Descriptor: | Complement factor B, SULFATE ION, ZINC ION, ... | | Authors: | Adams, C.M, Sellner, H, Ehara, T, Mac Sweeney, A, Crowley, M, Anderson, K, Karki, R, Mainolfi, N, Valeur, E, Sirockin, F, Gerhartz, B, Erbel, P, Hughes, N, Smith, T.M, Cumin, F, Argikar, U, Mogi, M, Sedrani, R, Wiesmann, C, Jaffee, B, Maibaum, J, Flohr, S, Harrison, R, Eder, J. | | Deposit date: | 2019-02-22 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Small-molecule factor B inhibitor for the treatment of complement-mediated diseases.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6LCB
 
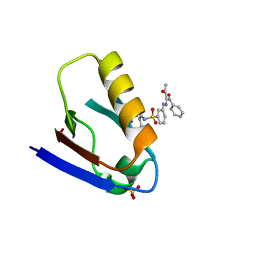 | | Crystal structure of human Dishevelled1 PDZ domain with its inhibitor NPL3009 | | Descriptor: | 2-[[3-[(2E)-2-[1,3-bis(oxidanylidene)-1-phenyl-butan-2-ylidene]hydrazinyl]phenyl]sulfonylamino]benzoic acid, SULFATE ION, Segment polarity protein dishevelled homolog DVL-1 | | Authors: | Yasukochi, S, Numoto, N, Tenno, N, Tenno, T, Ito, N, Hiroaki, H. | | Deposit date: | 2019-11-18 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of human Dishevelled1 PDZ domain with its inhibitor NPL3009
To Be Published
|
|
5GTG
 
 | | Crystal structure of onion lachrymatory factor synthase (LFS) containing 1,2-propanediol | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lachrymatory-factor synthase, S-1,2-PROPANEDIOL, ... | | Authors: | Arakawa, T, Sato, Y, Takabe, J, Masamura, N, Tsuge, N, Imai, S, Fushinobu, S. | | Deposit date: | 2016-08-20 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dissecting the Stereocontrolled Conversion of Short-Lived Sulfenic Acid by Lachrymatory Factor Synthase
Acs Catalysis, 10, 2020
|
|
5H4P
 
 | | Structural snapshot of cytoplasmic pre-60S ribosomal particles bound with Nmd3, Lsg1, Tif6 and Reh1 | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Ma, C, Wu, S, Li, N, Chen, Y, Yan, K, Li, Z, Zheng, L, Lei, J, Woolford, J.L, Gao, N. | | Deposit date: | 2016-11-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1
Nat. Struct. Mol. Biol., 24, 2017
|
|
4OAQ
 
 | | Crystal structure of the R-specific Carbonyl Reductase from Candida parapsilosis ATCC 7330 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Aggrawal, N, Mandal, P.K, Gautham, N, Chadha, A. | | Deposit date: | 2014-01-06 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.858 Å) | | Cite: | Insights into the stereoselectivity of R-specific Carbonyl Reductase from Candida parapsilosis ATCC 7330: Biochemical Characterization and Crystal structure studies
To be Published
|
|
5GPH
 
 | |
