6DW5
 
 | | SAMHD1 Bound to Gemcitabine-TP in the Catalytic Pocket | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-deoxy-2',2'-difluorocytidine 5'-(tetrahydrogen triphosphate), Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Knecht, K.M, Buzovetsky, O, Schneider, C, Thomas, D, Srikanth, V, Kaderali, L, Tofoleanu, F, Reiss, K, Ferreiros, N, Geisslinger, G, Batista, V.S, Ji, X, Cinatl, J, Keppler, O.T, Xiong, Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The structural basis for cancer drug interactions with the catalytic and allosteric sites of SAMHD1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DR0
 
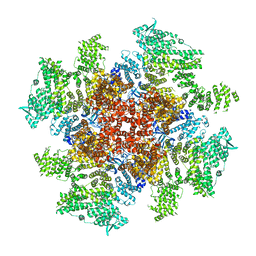 | | Class 5 IP3-bound human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2018-08-15 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DQ3
 
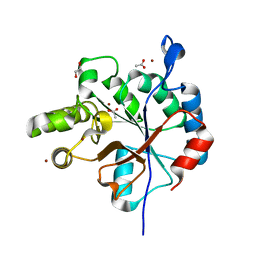 | | Streptococcus pyogenes deacetylase PplD in complex with acetate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, IMIDAZOLE, ... | | Authors: | Li, J, Korotkova, N, Korotkov, K.V. | | Deposit date: | 2018-06-10 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | PplD is a de-N-acetylase of the cell wall linkage unit of streptococcal rhamnopolysaccharides
Nat Commun, 13, 2022
|
|
6DQS
 
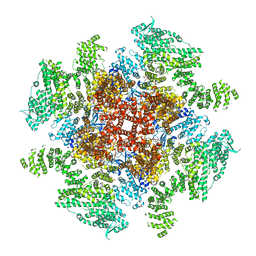 | | Class 3 IP3-bound human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DUV
 
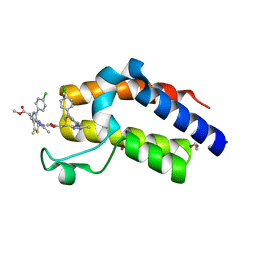 | | Crystal structure of the second bromodomain of human BRD4 in complex with MS417 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, methyl [(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetate | | Authors: | Babault, N, Zhou, M.M. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of Bound Water Molecules in Differential Recognition of Di-Acetylated Histone Peptides by the BRD4 Bromodomains
To be published
|
|
6DXK
 
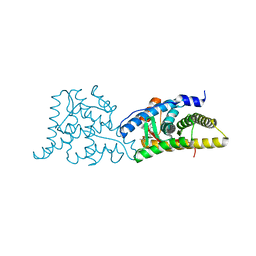 | | Glucocorticoid Receptor in complex with Compound 11 | | Descriptor: | (8S,11R,13S,14S,17S)-11-[4-(dimethylamino)phenyl]-17-(3,3-dimethylbut-1-yn-1-yl)-17-hydroxy-13-methyl-1,2,6,7,8,11,12,13,14,15,16,17-dodecahydro-3H-cyclopenta[a]phenanthren-3-one (non-preferred name), Glucocorticoid receptor | | Authors: | Rew, Y, Du, X, Eksterowicz, J, Zhou, H, Jahchan, N, Zhu, L, Yan, X, Kawai, H, McGee, L.R, Medina, J.C, Huang, T, Chen, C, Zavorotinskaya, T, Sutimantanapi, D, Waszczuk, J, Jackson, E, Huang, E, Ye, Q, Fantin, V.R, Daqing, S. | | Deposit date: | 2018-06-29 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Discovery of a Potent and Selective Steroidal Glucocorticoid Receptor Antagonist (ORIC-101).
J. Med. Chem., 61, 2018
|
|
1XAB
 
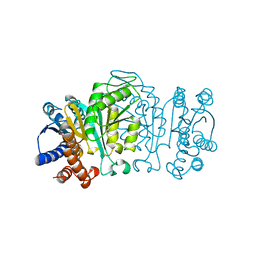 | |
1XAA
 
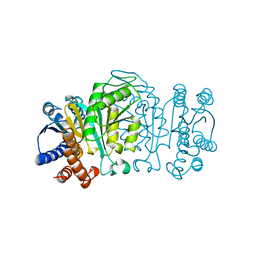 | |
6E15
 
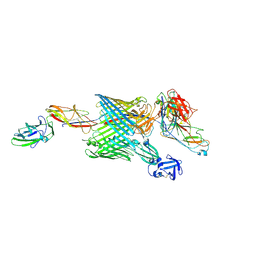 | | Handover mechanism of the growing pilus by the bacterial outer membrane usher FimD | | Descriptor: | Chaperone protein FimC, Fimbrial biogenesis outer membrane usher protein, Protein FimF, ... | | Authors: | Du, M, Yuan, Z, Yu, H, Henderson, N, Sarowar, S, Zhao, G, Werneburg, G.T, Thanassi, D.G, Li, H. | | Deposit date: | 2018-07-09 | | Release date: | 2018-10-17 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Handover mechanism of the growing pilus by the bacterial outer-membrane usher FimD.
Nature, 562, 2018
|
|
4M3F
 
 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | HTH-type transcriptional regulator EthR, N-(3-methylbutyl)-4-(2-methyl-1,3-thiazol-4-yl)benzenesulfonamide | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
6E14
 
 | | Handover mechanism of the growing pilus by the bacterial outer membrane usher FimD | | Descriptor: | Chaperone protein FimC, Fimbrial biogenesis outer membrane usher protein, Protein FimF, ... | | Authors: | Du, M, Yuan, Z, Yu, H, Henderson, N, Sarowar, S, Zhao, G, Werneburg, G.T, Thanassi, D.G, Li, H. | | Deposit date: | 2018-07-09 | | Release date: | 2018-10-17 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Handover mechanism of the growing pilus by the bacterial outer-membrane usher FimD.
Nature, 562, 2018
|
|
2K8T
 
 | | Solution NMR structure of trans-4-hydroxynonenal derived dG adduct of (6R,8S,11R)-configuration opposite dC | | Descriptor: | (2S,5R)-5-pentyltetrahydrofuran-2-ol, 5'-D(*DGP*DCP*DTP*DAP*DGP*DCP*DGP*DAP*DGP*DTP*DCP*DC)-3', 5'-D(*DGP*DGP*DAP*DCP*DTP*DCP*DGP*DCP*DTP*DAP*DGP*DC)-3' | | Authors: | Huang, H, Wang, H, Qi, N, Lloyd, R.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2008-09-23 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The stereochemistry of trans-4-hydroxynonenal-derived exocyclic 1,N2-2'-deoxyguanosine adducts modulates formation of interstrand cross-links in the 5'-CpG-3' sequence.
Biochemistry, 47, 2008
|
|
6EE3
 
 | | X-ray crystal structure of Pf-M1 in complex with inhibitor (6k) and catalytic zinc ion | | Descriptor: | (1R,2r,3S,5R,7R)-N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)ethyl]tricyclo[3.3.1.1~3,7~]decane-2-carboxamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2018-08-13 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Hydroxamic Acid Inhibitors Provide Cross-Species Inhibition of Plasmodium M1 and M17 Aminopeptidases.
J. Med. Chem., 62, 2019
|
|
6EED
 
 | | X-ray crystal structure of Pf-M1 in complex with inhibitor (6p) and catalytic zinc ion | | Descriptor: | (2R)-2-[(cyclohexylacetyl)amino]-N-hydroxy-2-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)acetamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2018-08-13 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hydroxamic Acid Inhibitors Provide Cross-Species Inhibition of Plasmodium M1 and M17 Aminopeptidases.
J. Med. Chem., 62, 2019
|
|
6E36
 
 | |
6E3F
 
 | |
6E53
 
 | | Structure of TERT in complex with a novel telomerase inhibitor | | Descriptor: | MAGNESIUM ION, RNA/DNA hairpin, Telomerase reverse transcriptase, ... | | Authors: | Hernandez-Sanchez, W, Huang, W, Plucinsky, B, Garcia-Vazquez, N, Berdis, A.J, Skordalakes, E, Taylor, D.J. | | Deposit date: | 2018-07-19 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A non-natural nucleotide uses a specific pocket to selectively inhibit telomerase activity.
Plos Biol., 17, 2019
|
|
2MHZ
 
 | | Structure of Exocyclic S,S N6,N6-(2,3-Dihydroxy-1,4-butadiyl)-2'-Deoxyadenosine Adduct Induced by 1,2,3,4-Diepoxybutane in DNA | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(SDE)P*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3' | | Authors: | Kowal, E.A, Seneviratne, U, Wickramaratne, S, Doherty, K.E, Cao, X, Tretyakova, N, Stone, M.P. | | Deposit date: | 2013-12-05 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of Exocyclic R,R- and S,S-N(6),N(6)-(2,3-Dihydroxybutan-1,4-diyl)-2'-Deoxyadenosine Adducts Induced by 1,2,3,4-Diepoxybutane.
Chem.Res.Toxicol., 27, 2014
|
|
2M0G
 
 | | Structure, phosphorylation and U2AF65 binding of the Nterminal Domain of splicing factor 1 during 3 splice site Recognition | | Descriptor: | Splicing factor 1, Splicing factor U2AF 65 kDa subunit | | Authors: | Madl, T, Sattler, M, Zhang, Y, Bagdiul, I, Kern, T, Kang, H, Zou, P, Maeusbacher, N, Sieber, S.A, Kraemer, A. | | Deposit date: | 2012-10-25 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition.
Nucleic Acids Res., 41, 2013
|
|
2MHX
 
 | | Structure of Exocyclic R,R N6,N6-(2,3-Dihydroxy-1,4-butadiyl)-2'-Deoxyadenosine Adduct Induced by 1,2,3,4-Diepoxybutane in DNA | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(RBD)P*AP*GP*AP*AP*G)-3'), 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3') | | Authors: | Kowal, E.A, Seneviratne, U, Wickramaratne, S, Doherty, K.E, Cao, X, Tretyakova, N, Stone, M.P. | | Deposit date: | 2013-12-05 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of Exocyclic R,R- and S,S-N(6),N(6)-(2,3-Dihydroxybutan-1,4-diyl)-2'-Deoxyadenosine Adducts Induced by 1,2,3,4-Diepoxybutane.
Chem.Res.Toxicol., 27, 2014
|
|
2MNX
 
 | | Major groove orientation of the (2S)-N6-(2-hydroxy-3-buten-1-yl)-2'-deoxyadenosine DNA adduct induced by 1,2-epoxy-3-butene | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(6HB)P*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3' | | Authors: | Kowal, E.A, Kotapati, S, Turo, M, Tretyakova, N, Stone, M.P. | | Deposit date: | 2014-04-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Major Groove Orientation of the (2S)-N(6)-(2-Hydroxy-3-buten-1-yl)-2'-deoxyadenosine DNA Adduct Induced by 1,2-Epoxy-3-butene.
Chem.Res.Toxicol., 27, 2014
|
|
4QOS
 
 | | CRYSTAL STRUCTURE OF PSPF(1-265) E108Q MUTANT bound to ADP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | Deposit date: | 2014-06-20 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
4QNR
 
 | | CRYSTAL STRUCTURE OF PSPF(1-265) E108Q MUTANT bound to ATP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
2L62
 
 | | Protein and metal cluster structure of the wheat metallothionein domain g-Ec-1. The second part of the puzzle. | | Descriptor: | EC protein I/II, ZINC ION | | Authors: | Loebus, J, Peroza, E.A, Bluethgen, N, Fox, T, Meyer-Klaucke, W, Zerbe, O, Freisinger, E. | | Deposit date: | 2010-11-12 | | Release date: | 2011-05-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Protein and metal cluster structure of the wheat metallothionein domain gamma-E(c)-1: the second part of the puzzle.
J.Biol.Inorg.Chem., 16, 2011
|
|
4QNM
 
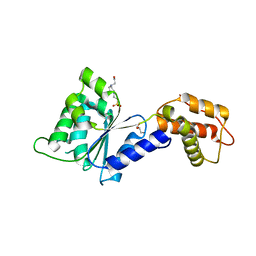 | | CRYSTAL STRUCTURE of PSPF(1-265) E108Q MUTANT | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Psp operon transcriptional activator | | Authors: | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.628 Å) | | Cite: | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
