3C0F
 
 | | Crystal Structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source | | Descriptor: | Uncharacterized protein AF_1514 | | Authors: | Li, Y, Bahti, P, Shaw, N, Song, G, Yin, J, Zhu, J.-Y, Zhang, H, Xu, H, Wang, B.-C, Liu, Z.-J. | | Deposit date: | 2008-01-20 | | Release date: | 2008-02-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source.
Proteins, 71, 2008
|
|
6ENK
 
 | | The X-ray crystal structure of DesE bound to desferrioxamine B | | Descriptor: | DesE, SODIUM ION, desferrioxamine B | | Authors: | Naismith, J.H, McMahon, S.A, Challis, G.L, Kadi, N, Oke, M, Liu, H, Carter, L.G, Johnson, K.A. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Desferrioxamine biosynthesis: diverse hydroxamate assembly by substrate-tolerant acyl transferase DesC.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
3BXI
 
 | | Structure of the complex of bovine lactoperoxidase with its catalyzed product hypothiocyanate ion at 2.3A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, A.K, Singh, N, Sharma, S, Shin, K, Takase, M, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2008-01-14 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibition of lactoperoxidase by its own catalytic product: crystal structure of the hypothiocyanate-inhibited bovine lactoperoxidase at 2.3-A resolution.
Biophys.J., 96, 2009
|
|
3C5A
 
 | | Crystal structure of the C-terminal deleted mutant of the class A carbapenemase KPC-2 at 1.23 angstrom | | Descriptor: | CITRIC ACID, Class A carbapenemase KPC-2 | | Authors: | Petrella, S, Ziental-Gelus, N, Mayer, C, Jarlier, V, Sougakoff, W. | | Deposit date: | 2008-01-31 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Genetic and structural insights into the dissemination potential of the extremely-broad-spectrum class A {beta}-lactamase (EBSBL) KPC-2 identified in two strains of Escherichia coli and Enterobacter cloacae isolated from the same patient in France
ANTIMICROB.AGENTS CHEMOTHER., 2008
|
|
6ER4
 
 | | Ruminococcus gnavus IT-sialidase CBM40 bound to alpha2,6 sialyllactose | | Descriptor: | BNR/Asp-box repeat protein, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2017-10-16 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Unravelling the specificity and mechanism of sialic acid recognition by the gut symbiont Ruminococcus gnavus.
Nat Commun, 8, 2017
|
|
3C8H
 
 | | Crystal structure of the enterobactin esterase FES from Shigella flexneri in the presence of 2,3-Di-hydroxy-N-benzoyl-serine | | Descriptor: | Enterochelin esterase | | Authors: | Kim, Y, Maltseva, N, Abergel, R, Holzle, D, Raymond, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-12 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Siderophore Mediated Iron Acquisition: Structure and Specificity of Enterobactin Esterase from Shigella flexneri.
To be Published
|
|
6EE2
 
 | |
3C05
 
 | | Crystal structure of Acostatin from Agkistrodon Contortrix Contortrix | | Descriptor: | Disintegrin acostatin alpha, Disintegrin acostatin-beta, SULFATE ION | | Authors: | Moiseeva, N, Bau, R, Allaire, M. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of acostatin, a dimeric disintegrin from southern copperhead
(Agkistrodon contortrix contortrix) at 1.7 A resolution
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3C1T
 
 | | Binding of two substrate analogue molecules to dihydroflavonol 4-reductase alters the functional geometry of the catalytic site | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, dihydroflavonol 4-reductase | | Authors: | Trabelsi, N, Petit, P, Granier, T, Langlois d'Estaintot, B, Delrot, S, Gallois, B. | | Deposit date: | 2008-01-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Structural evidence for the inhibition of grape dihydroflavonol 4-reductase by flavonols
Acta Crystallogr.,Sect.D, D64, 2008
|
|
6E8G
 
 | |
3BLP
 
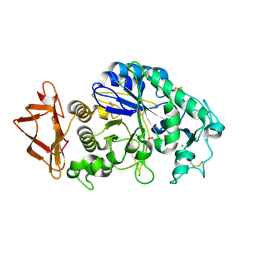 | | Role of aromatic residues in human salivary alpha-amylase | | Descriptor: | 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 5-HYDROXYMETHYL-CHONDURITOL, Alpha-amylase 1, ... | | Authors: | Ramasubbu, N. | | Deposit date: | 2007-12-11 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-function relationships in human salivary alpha-amylase: role of aromatic residues in a secondary binding site
Biologia, 63, 2008
|
|
3BMV
 
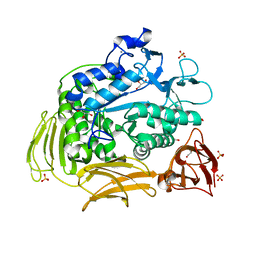 | | Cyclodextrin glycosyl transferase from Thermoanerobacterium thermosulfurigenes EM1 mutant S77P | | Descriptor: | CALCIUM ION, Cyclomaltodextrin glucanotransferase, GLYCEROL, ... | | Authors: | Rozeboom, H.J, van Oosterwijk, N, Dijkstra, B.W. | | Deposit date: | 2007-12-13 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Elimination of competing hydrolysis and coupling side reactions of a cyclodextrin glucanotransferase by directed evolution.
Biochem.J., 413, 2008
|
|
3BXA
 
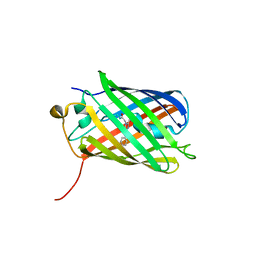 | |
3BYI
 
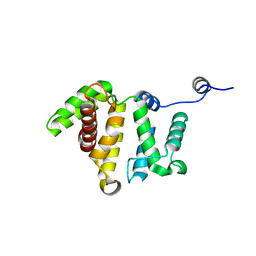 | | Crystal structure of human Rho GTPase activating protein 15 (ARHGAP15) | | Descriptor: | Rho GTPase activating protein 15 | | Authors: | Shrestha, L, Tickle, J, Elkins, J, Burgess-Brown, N, Johansson, C, Papagrigoriou, E, Kavanagh, K, Pike, A.C.W, Ugochukwu, E, Uppenberg, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Human Rho GTPase Activating Protein 15 (ARHGAP15).
To be Published
|
|
3BSH
 
 | |
6ER2
 
 | |
6ECE
 
 | | Vlm2 thioesterase domain with genetically encoded 2,3-diaminopropionic acid bound with a dodecadepsipeptide, space group H3 | | Descriptor: | Vlm2, dodecadepsipeptide | | Authors: | Alonzo, D.A, Huguenin-Dezot, N, Heberlig, G.W, Mahesh, M, Nguyen, D.P, Dornan, M.H, Boddy, C.N, Chin, J.W, Schmeing, T.M. | | Deposit date: | 2018-08-07 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trapping biosynthetic acyl-enzyme intermediates with encoded 2,3-diaminopropionic acid.
Nature, 565, 2019
|
|
6EE6
 
 | | X-ray crystal structure of Pf-M1 in complex with inhibitor (6o) and catalytic zinc ion | | Descriptor: | (2R)-2-[(cyclopentylacetyl)amino]-N-hydroxy-2-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)acetamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2018-08-13 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hydroxamic Acid Inhibitors Provide Cross-Species Inhibition of Plasmodium M1 and M17 Aminopeptidases.
J. Med. Chem., 62, 2019
|
|
4Y2B
 
 | | Co-crystal structure of 3-ethyl-2-(isopropylamino)-7-(pyridin-3-yl)thieno[3,2-d]pyrimidin-4(3H)-one bound to PDE7A | | Descriptor: | 3-ethyl-2-(propan-2-ylamino)-7-(pyridin-3-yl)thieno[3,2-d]pyrimidin-4(3H)-one, High affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A, MAGNESIUM ION, ... | | Authors: | Endo, Y, Kawai, K, Asano, T, Amano, S, Asanuma, Y, Sawada, K, Onodera, Y, Ueo, N, Takahashi, N, Sonoda, Y, Kamei, N, Irie, T. | | Deposit date: | 2015-02-09 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2-(Isopropylamino)thieno[3,2-d]pyrimidin-4(3H)-one derivatives as selective phosphodiesterase 7 inhibitors with potent in vivo efficacy
Bioorg.Med.Chem.Lett., 25, 2015
|
|
2AC0
 
 | | Structural Basis of DNA Recognition by p53 Tetramers (complex I) | | Descriptor: | 5'-D(*CP*GP*GP*GP*CP*AP*TP*GP*CP*CP*CP*G)-3', Cellular tumor antigen p53, ZINC ION | | Authors: | Kitayner, M, Rozenberg, H, Kessler, N, Rabinovich, D, Shakked, Z. | | Deposit date: | 2005-07-18 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of DNA Recognition by p53 Tetramers
Mol.Cell, 22, 2006
|
|
2AEM
 
 | | Crystal Structures of the MthK RCK Domain | | Descriptor: | Calcium-gated potassium channel mthK | | Authors: | Dong, J, Shi, N, Berke, I, Chen, L, Jiang, Y. | | Deposit date: | 2005-07-22 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the MthK RCK Domain and the Effect of Ca2+ on Gating Ring Stability
J.Biol.Chem., 280, 2005
|
|
9EZ1
 
 | | Vitamin D receptor in complex with 1,4a,25-trihydroxyvitamin D3 | | Descriptor: | 1,4a,25-trihydroxyvitamin D3, ACETATE ION, Nuclear receptor coactivator 2, ... | | Authors: | Rochel, N. | | Deposit date: | 2024-04-10 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 4-Hydroxy-1 alpha ,25-Dihydroxyvitamin D 3 : Synthesis and Structure-Function Study.
Biomolecules, 14, 2024
|
|
8Y4U
 
 | | Crystal structure of a His1 from oryza sativa | | Descriptor: | FE (III) ION, Fe(II)/2-oxoglutarate-dependent oxygenase | | Authors: | Wang, N, Ma, J.M, Shibing, H, Beibei, Y, He, Z, Dandan, L. | | Deposit date: | 2024-01-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of HPPD inhibitor sensitive protein from Oryza sativa.
Biochem.Biophys.Res.Commun., 704, 2024
|
|
1ZXK
 
 | | Crystal Structure of Cadherin8 EC1 domain | | Descriptor: | Cadherin-8 | | Authors: | Patel, S.D, Ciatto, C, Chen, C.P, Bahna, F, Arkus, N, Schieren, I, Rajebhosale, M, Jessell, T.M, Honig, B, Price, S.R, Shapiro, L. | | Deposit date: | 2005-06-08 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Type II cadherin ectodomain structures: implications for classical cadherin specificity.
Cell(Cambridge,Mass.), 124, 2006
|
|
9FQ9
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalently bound inhibitor PSB-21110 (compound 29b in publication) | | Descriptor: | (5-chloranylpyridin-3-yl) 4-ethoxy-2-fluoranyl-benzoate, BROMIDE ION, Non-structural protein 11 | | Authors: | Strater, N, Claff, T, Sylvester, K, Oneto, A, Guetschow, M, Mueller, C.E. | | Deposit date: | 2024-06-19 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Nonpeptidic Irreversible Inhibitors of SARS-CoV-2 Main Protease with Potent Antiviral Activity.
J.Med.Chem., 67, 2024
|
|
