6XJ3
 
 | | Crystal structure of Class D beta-lactamase from Klebsiella quasipneumoniae in complex with avibactam | | 分子名称: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-22 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Class D beta-lactamase from Klebsiella quasipneumoniae
To Be Published
|
|
5WR5
 
 | | Thermolysin, liganded form with cryo condition 1 | | 分子名称: | CALCIUM ION, N-[(benzyloxy)carbonyl]-L-aspartic acid, TETRAETHYLENE GLYCOL, ... | | 著者 | Kunishima, N, Naitow, H, Matsuura, Y. | | 登録日 | 2016-11-29 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Protein-ligand complex structure from serial femtosecond crystallography using soaked thermolysin microcrystals and comparison with structures from synchrotron radiation
Acta Crystallogr D Struct Biol, 73, 2017
|
|
8RJY
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalent inhibitor GUE-3899 (compound 58 in publication) | | 分子名称: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-1-[[(2~{S})-1-[[(4-chlorophenyl)methyl-(iminomethyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]thiophene-2-carboxamide | | 著者 | Strater, N, Claff, T, Sylvester, K, Mueller, C.E, Guetschow, M, Useini, A. | | 登録日 | 2023-12-22 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Macrocyclic Azapeptide Nitriles: Structure-Based Discovery of Potent SARS-CoV-2 Main Protease Inhibitors as Antiviral Drugs.
J.Med.Chem., 67, 2024
|
|
8RSB
 
 | | p97 (VCP) mutant - F539A ADP state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | 著者 | Arie, M, Matzov, D, Karmona, R, Szenkier, N, Stanhill, A, Navon, A. | | 登録日 | 2024-01-24 | | 公開日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | p97 (VCP) mutant - F539A ADP state
To Be Published
|
|
8RJV
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalent inhibitor GUE-3778 (compound 12 in publication) | | 分子名称: | (phenylmethyl) ~{N}-[(2~{S})-1-[[(3-chloranyl-2-fluoranyl-phenyl)methyl-(iminomethyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]carbamate, 3C-like proteinase nsp5 | | 著者 | Strater, N, Claff, T, Sylvester, K, Mueller, C.E, Guetschow, M, Useini, A. | | 登録日 | 2023-12-21 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Macrocyclic Azapeptide Nitriles: Structure-Based Discovery of Potent SARS-CoV-2 Main Protease Inhibitors as Antiviral Drugs.
J.Med.Chem., 67, 2024
|
|
8RJZ
 
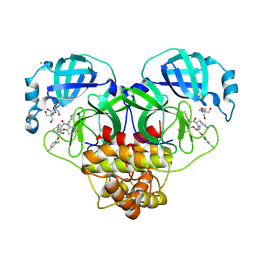 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GUE-3801 (compound 80 in publication) | | 分子名称: | (7~{S})-6-[2-[2,4-bis(chloranyl)phenoxy]ethanoyl]-14-fluoranyl-10-(iminomethyl)-9-methyl-7-(phenylmethyl)-2-oxa-6,9,10-triazabicyclo[10.4.0]hexadeca-1(12),13,15-trien-8-one, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase nsp5, ... | | 著者 | Strater, N, Claff, T, Sylvester, K, Mueller, C.E, Guetschow, M, Useini, A. | | 登録日 | 2023-12-22 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Macrocyclic Azapeptide Nitriles: Structure-Based Discovery of Potent SARS-CoV-2 Main Protease Inhibitors as Antiviral Drugs.
J.Med.Chem., 67, 2024
|
|
8RSC
 
 | | p97 (VCP) mutant - F539A | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | 著者 | Arie, M, Matzov, D, Karmona, R, Szenkier, N, Stanhill, A, Navon, A. | | 登録日 | 2024-01-24 | | 公開日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | p97 (VCP) mutant - F539A
To Be Published
|
|
6XKX
 
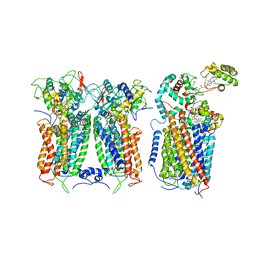 | | R. capsulatus CIII2CIV tripartite super-complex, conformation A (SC-1A) | | 分子名称: | COPPER (II) ION, Cbb3-type cytochrome c oxidase subunit CcoP,Cytochrome c-type cyt cy, Cytochrome b, ... | | 著者 | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | 登録日 | 2020-06-27 | | 公開日 | 2020-12-30 | | 最終更新日 | 2021-03-03 | | 実験手法 | ELECTRON MICROSCOPY (6.1 Å) | | 主引用文献 | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
6GC5
 
 | |
5WV7
 
 | |
6GAZ
 
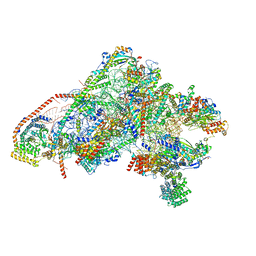 | | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. This file contains the 28S ribosomal subunit. | | 分子名称: | 12S ribosomal RNA, mitochondrial, 28S ribosomal protein S18b, ... | | 著者 | Kummer, E, Leibundgut, M, Boehringer, D, Ban, N. | | 登録日 | 2018-04-13 | | 公開日 | 2018-08-08 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM.
Nature, 560, 2018
|
|
5X0N
 
 | |
6XMJ
 
 | | Human 20S proteasome bound to an engineered 11S (PA26) activator | | 分子名称: | Proteasome activator protein PA26, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | 著者 | de la Pena, A.H, Opoku-Nsiah, K.A, Williams, S.K, Chopra, N, Sali, A, Gestwicki, J.E, Lander, G.C. | | 登録日 | 2020-06-30 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | The Y Phi motif defines the structure-activity relationships of human 20S proteasome activators.
Nat Commun, 13, 2022
|
|
5WZJ
 
 | | Structure of APUM23-GGAUUUGACGG | | 分子名称: | Pumilio homolog 23, RNA (5'-R(*GP*GP*AP*UP*UP*UP*GP*AP*CP*GP*G)-3') | | 著者 | Bao, H, Wang, N, Wang, C, Jiang, Y, Wu, J, Shi, Y. | | 登録日 | 2017-01-18 | | 公開日 | 2017-09-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | Structural basis for the specific recognition of 18S rRNA by APUM23.
Nucleic Acids Res., 45, 2017
|
|
6XTK
 
 | | Y114C Transthyretin structure in complex with Tolcalpone | | 分子名称: | Tolcapone, Transthyretin | | 著者 | Varejao, N, Reverter, D, Pinheiro, F, Pallares, I, Ventura, S. | | 登録日 | 2020-01-16 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Tolcapone, a potent aggregation inhibitor for the treatment of familial leptomeningeal amyloidosis.
Febs J., 288, 2021
|
|
5X15
 
 | |
6XUG
 
 | |
6XUQ
 
 | |
5V9I
 
 | | Crystal structure of catalytic domain of G9a with MS0105 | | 分子名称: | GLYCEROL, Histone-lysine N-methyltransferase EHMT2, N~2~-cyclohexyl-N~4~-(1-ethylpiperidin-4-yl)-6,7-dimethoxy-N~2~-methylquinazoline-2,4-diamine, ... | | 著者 | Dong, A, Zeng, H, Liu, J, Xiong, Y, Babault, N, Jin, J, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Wu, H, Brown, P.J, Structural Genomics Consortium (SGC) | | 登録日 | 2017-03-23 | | 公開日 | 2018-03-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Crystal structure of catalytic domain of G9a with MS0105
to be published
|
|
6XXN
 
 | | Crystal structure of NB7, a nanobody targeting prostate specific membrane antigen | | 分子名称: | NB_7_a,b,c,f, NB_7_g, NB_7_h, ... | | 著者 | Shahar, A, Rosenfeld, L, Papo, N. | | 登録日 | 2020-01-28 | | 公開日 | 2020-06-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Nanobodies Targeting Prostate-Specific Membrane Antigen for the Imaging and Therapy of Prostate Cancer.
J.Med.Chem., 63, 2020
|
|
6G8H
 
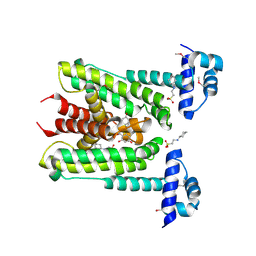 | | Flavonoid-responsive Regulator FrrA in complex with (R,S)-Naringenin | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, NARINGENIN, R-naringenin, ... | | 著者 | Werner, N, Hoppen, J, Palm, G, Werten, S, Goettfert, M, Hinrichs, W. | | 登録日 | 2018-04-08 | | 公開日 | 2019-04-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The induction mechanism of the flavonoid-responsive regulator FrrA.
Febs J., 2021
|
|
8TL8
 
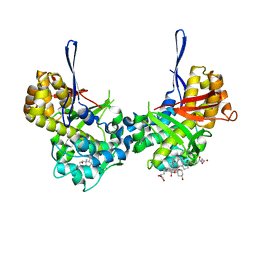 | | Structure of Orthoreovirus RNA Chaperone SigmaNS R6A mutant in complex with bile acid | | 分子名称: | GLYCOCHOLIC ACID, Protein sigma-NS | | 著者 | Prasad, B.V.V, Zhao, B, Hu, L, Neetu, N. | | 登録日 | 2023-07-26 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure of orthoreovirus RNA chaperone sigma NS, a component of viral replication factories.
Nat Commun, 15, 2024
|
|
5UK0
 
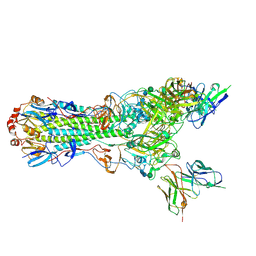 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | 著者 | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | 登録日 | 2017-01-19 | | 公開日 | 2017-05-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
6X6L
 
 | | Cryo-EM Structure of CagX and CagY within the dCag3 Helicobacter pylori PR | | 分子名称: | Cag pathogenicity island protein (Cag7), Cag pathogenicity island protein (Cag8) | | 著者 | Sheedlo, M.J, Chung, J.M, Sawhney, N, Durie, C.L, Cover, T.L, Ohi, M.D, Lacy, D.B. | | 登録日 | 2020-05-28 | | 公開日 | 2020-09-30 | | 最終更新日 | 2021-04-14 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Cryo-EM reveals species-specific components within the Helicobacter pylori Cag type IV secretion system core complex.
Elife, 9, 2020
|
|
5UML
 
 | |
