5WCR
 
 | | Phosphotriesterase variant R0deltaL7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R0, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-07-01 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Phosphotriesterase variant R0deltaL7
To Be Published
|
|
5WDA
 
 | | Structure of the PulG pseudopilus | | Descriptor: | CALCIUM ION, General secretion pathway protein G | | Authors: | Lopez-Castilla, A, Thomassin, J.L, Bardiaux, B, Zheng, W, Nivaskumar, M, Yu, X, Nilges, M, Egelman, E.H, Izadi-Pruneyre, N, Francetic, O. | | Deposit date: | 2017-07-04 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structure of the calcium-dependent type 2 secretion pseudopilus.
Nat Microbiol, 2, 2017
|
|
5WF6
 
 | | Agonist bound human A2a adenosine receptor with S91A mutation at 2.90 A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 6-(2,2-diphenylethylamino)-9-[(2R,3R,4S,5S)-5-(ethylcarbamoyl)-3,4-dihydroxy-oxolan-2-yl]-N-[2-[(1-pyridin-2-ylpiperidin-4-yl)carbamoylamino]ethyl]purine-2-carboxamide, Human A2a adenosine receptor T4L chimera | | Authors: | White, K.L, Eddy, M.T, Gao, Z, Han, G.W, Hanson, M.A, Lian, T, Deary, A, Patel, N, Jacobson, K.A, Katritch, V, Stevens, R.C. | | Deposit date: | 2017-07-11 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Connection between Activation Microswitch and Allosteric Sodium Site in GPCR Signaling.
Structure, 26, 2018
|
|
5WWK
 
 | | Highly stable green fluorescent protein | | Descriptor: | Green fluorescent protein | | Authors: | Sriram, R, George, A, Kesavan, M, Jaimohan, S.M, Kamini, N.R, Easwaramoorthi, S, Ganesh, S, Gunasekaran, K, Ayyadurai, N. | | Deposit date: | 2017-01-02 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Excited State Electronic Interconversion and Structural Transformation of Engineered Red-Emitting Green Fluorescent Protein Mutant.
J.Phys.Chem.B, 123, 2019
|
|
8DN8
 
 | |
5WZH
 
 | | Structure of APUM23-GGAAUUGACGG | | Descriptor: | Pumilio homolog 23, RNA (5'-R(*GP*GP*AP*AP*UP*UP*GP*AP*CP*GP*G)-3') | | Authors: | Bao, H, Wang, N, Wang, C, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-01-18 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Structural basis for the specific recognition of 18S rRNA by APUM23.
Nucleic Acids Res., 45, 2017
|
|
8DKU
 
 | | CryoEM structure of the A. aeolicus WzmWzt transporter bound to the native O antigen | | Descriptor: | 6-deoxy-3-O-methyl-alpha-D-mannopyranose-(1-3)-beta-D-rhamnopyranose-(1-2)-alpha-D-rhamnopyranose, ABC transporter, Transport permease protein | | Authors: | Spellmon, N, Muszynski, A, Vlach, J, Zimmer, J. | | Deposit date: | 2022-07-06 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis for polysaccharide recognition and modulated ATP hydrolysis by the O antigen ABC transporter.
Nat Commun, 13, 2022
|
|
8DNC
 
 | | CryoEM structure of the A. aeolicus WzmWzt transporter bound to the native O antigen and ADP | | Descriptor: | 6-deoxy-3-O-methyl-alpha-D-mannopyranose-(1-3)-beta-D-rhamnopyranose-(1-2)-alpha-D-rhamnopyranose, ABC transporter, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Spellmon, N, Muszynski, A, Vlach, J, Zimmer, J. | | Deposit date: | 2022-07-11 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for polysaccharide recognition and modulated ATP hydrolysis by the O antigen ABC transporter.
Nat Commun, 13, 2022
|
|
5WIZ
 
 | | Phosphotriesterase variant S5 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-07-21 | | Release date: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Phosphotriesterase variant S5
To Be Published
|
|
8EBF
 
 | |
8E1A
 
 | | Structure-based study to overcome cross-reactivity of novel androgen receptor inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(3-fluoro-2-methoxyphenyl)-1,3-thiazol-2-yl]morpholine, Androgen receptor | | Authors: | Lallous, N, Li, H, Radaeva, M, Dalal, K, Leblanc, E, Ban, F, Ciesielski, F, Chow, B, Morin, M, Singh, K, Rennie, P.S, Cherkasov, A. | | Deposit date: | 2022-08-10 | | Release date: | 2022-09-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Based Study to Overcome Cross-Reactivity of Novel Androgen Receptor Inhibitors.
Cells, 11, 2022
|
|
5WKX
 
 | | Barium sites in the structure of a resting acid sensing ion channel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, ... | | Authors: | Yoder, N, Gouaux, E. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.034 Å) | | Cite: | Divalent cation and chloride ion sites of chicken acid sensing ion channel 1a elucidated by x-ray crystallography.
PLoS ONE, 13, 2018
|
|
8DL0
 
 | |
5X0O
 
 | |
8E1F
 
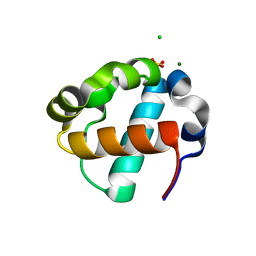 | | Sterile Alpha Motif Domain of Human Translocation ETS Leukemia, Non-Polymer Crystal Form | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Wilson, E, Nawarathnage, S, Stewart, C, Brown, S, Bezzant, B, Bunn, D, Towne, N, Pedroza Romo, M.J, Moody, J.D. | | Deposit date: | 2022-08-10 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Sterile Alpha Motif Domain of Human Translocation ETS Leukemia, Non-Polymer Crystal Form
To Be Published
|
|
5WIS
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with methymycin and bound to mRNA and A-, P- and E-site tRNAs at 2.7A resolution | | Descriptor: | (3R,4S,5S,7R,9E,11S,12R)-12-ethyl-11-hydroxy-3,5,7,11-tetramethyl-2,8-dioxooxacyclododec-9-en-4-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 16S Ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Almutairi, M.M, Svetlov, M.S, Hansen, D.A, Khabibullina, N.F, Klepacki, D, Kang, H.Y, Sherman, D.H, Vazquez-Laslop, N, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2017-07-20 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Co-produced natural ketolides methymycin and pikromycin inhibit bacterial growth by preventing synthesis of a limited number of proteins.
Nucleic Acids Res., 45, 2017
|
|
8DKY
 
 | |
8DNE
 
 | |
8DKS
 
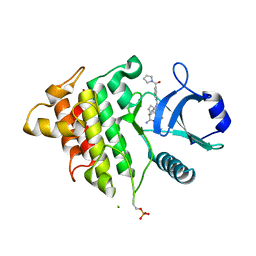 | | IRAK4 IN COMPLEX WITH COMPOUND #3 | | Descriptor: | (1S,2S,3R,4R)-3-({2-[3-(pyrrolidine-1-carbonyl)anilino]thieno[3,2-d]pyrimidin-4-yl}amino)bicyclo[2.2.1]hept-5-ene-2-carboxamide, BETA-MERCAPTOETHANOL, Interleukin-1 receptor-associated kinase 4 | | Authors: | Chen, Y, Lin, N. | | Deposit date: | 2022-07-06 | | Release date: | 2022-08-03 | | Last modified: | 2022-08-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Bicyclic pyrimidine compounds as potent IRAK4 inhibitors.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
5WRQ
 
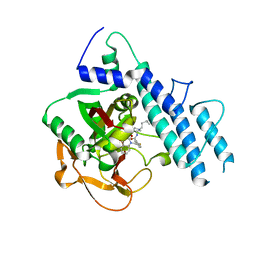 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 5-[[2,4-bis(oxidanylidene)quinazolin-1-yl]methyl]-2-fluoranyl-N-[(3R)-1-(3-methylbutyl)pyrrolidin-3-yl]benzamide, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | Deposit date: | 2016-12-03 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
5WIT
 
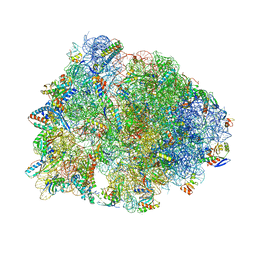 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with pikromycin and bound to mRNA and A-, P- and E-site tRNAs at 2.6A resolution | | Descriptor: | (3R,5R,6S,7S,9R,11E,13S,14R)-14-ethyl-13-hydroxy-3,5,7,9,13-pentamethyl-2,4,10-trioxo-1-oxacyclotetradec-11-en-6-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Almutairi, M.M, Svetlov, M.S, Hansen, D.A, Khabibullina, N.F, Klepacki, D, Kang, H.Y, Sherman, D.H, Vazquez-Laslop, N, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2017-07-20 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Co-produced natural ketolides methymycin and pikromycin inhibit bacterial growth by preventing synthesis of a limited number of proteins.
Nucleic Acids Res., 45, 2017
|
|
5WS0
 
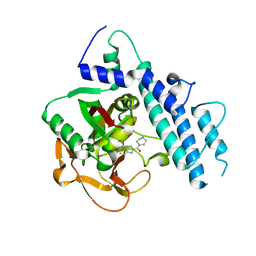 | | Structure of human PARP1 catalytic domain bound to a benzoimidazole inhibitor | | Descriptor: | 2-piperazin-1-ylcarbonyl-1H-benzimidazole-4-carboxamide, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a benzoimidazole inhibitor
To Be Published
|
|
5WSD
 
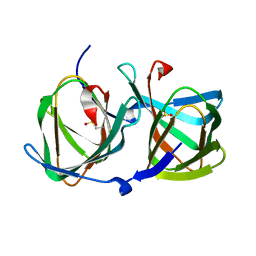 | | Crystal structure of a cupin protein (tm1459) in apo form | | Descriptor: | Uncharacterized protein tm1459 | | Authors: | Fujieda, N, Nakano, T, Taniguchi, Y, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2016-12-06 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Well-Defined Osmium-Cupin Complex: Hyperstable Artificial Osmium Peroxygenase
J. Am. Chem. Soc., 2017
|
|
5WR6
 
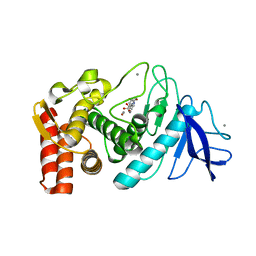 | | Thermolysin, liganded form with cryo condition 2 | | Descriptor: | CALCIUM ION, N-[(benzyloxy)carbonyl]-L-aspartic acid, Thermolysin, ... | | Authors: | Kunishima, N, Naitow, H, Matsuura, Y. | | Deposit date: | 2016-11-29 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein-ligand complex structure from serial femtosecond crystallography using soaked thermolysin microcrystals and comparison with structures from synchrotron radiation
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5WS1
 
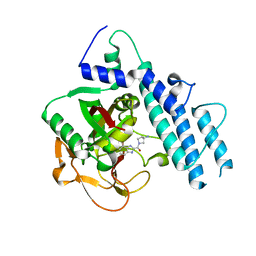 | | Structure of human PARP1 catalytic domain bound to a benzoimidazole inhibitor | | Descriptor: | 2-[(3R)-3-azanylpyrrolidin-1-yl]carbonyl-1H-benzimidazole-4-carboxamide, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a benzoimidazole inhibitor
To Be Published
|
|
