2ZCT
 
 | | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate | | Descriptor: | Probable peroxiredoxin | | Authors: | Nakamura, T, Hagihara, Y, Abe, M, Inoue, T, Yamamoto, T, Matsumura, H. | | Deposit date: | 2007-11-12 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1MBE
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBJ
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBG
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 2 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBK
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBF
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
8YHR
 
 | | DHODH in complex with furocoumavirin | | Descriptor: | 4-methyl-8-[(S)-oxidanyl(phenyl)methyl]-9-phenyl-furo[2,3-h]chromen-2-one, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Hara, K, Okumura, H, Nakahara, M, Sato, M, Hashimoto, H, Osada, H, Watanabe, K. | | Deposit date: | 2024-02-28 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Analyses of Inhibition of Human Dihydroorotate Dehydrogenase by Antiviral Furocoumavirin.
Biochemistry, 63, 2024
|
|
9ARQ
 
 | | Crystal structure of SARS-CoV-2 main protease (authentic protein) in complex with an inhibitor TKB-245 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
9ARS
 
 | | Crystal structure of SARS-CoV-2 main protease E166V mutant in complex with an inhibitor TKB-245 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Misumi, S, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
9ART
 
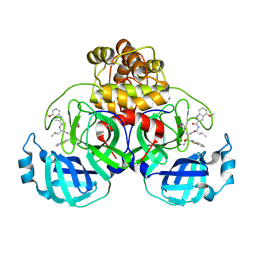 | | Crystal structure of SARS-CoV-2 main protease A191T mutant in complex with an inhibitor 5h | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
9AVQ
 
 | | Crystal structure of SARS-CoV-2 main protease A191T mutant in complex with an inhibitor Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-03-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
1EW0
 
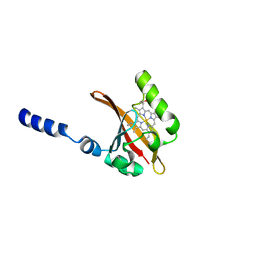 | | CRYSTAL STRUCTURE ANALYSIS OF THE SENSOR DOMAIN OF RMFIXL(FERROUS FORM) | | Descriptor: | FIXL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Miyatake, H, Mukai, M, Park, S.-Y, Adachi, S, Tamura, K, Nakamura, H, Nakamura, K, Tsuchiya, T, Iizuka, T, Shiro, Y. | | Deposit date: | 2000-04-21 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Sensory mechanism of oxygen sensor FixL from Rhizobium meliloti: crystallographic, mutagenesis and resonance Raman spectroscopic studies
J.MOL.BIOL., 301, 2000
|
|
1D06
 
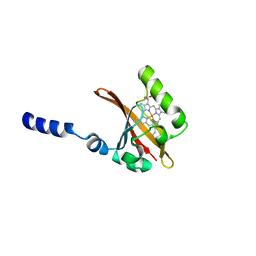 | | STRUCTURAL BASIS OF DIMERIZATION AND SENSORY MECHANISMS OF OXYGEN-SENSING DOMAIN OF RHIZOBIUM MELILOTI FIXL DETERMINED AT 1.4A RESOLUTION | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, nitrogen fixation regulatory protein fixL | | Authors: | Miyatake, H, Mukai, M, Park, S.-Y, Adachi, S, Tamura, K, Nakamura, H, Nakamura, K, Tsuchiya, T, Iizuka, T, Shiro, Y. | | Deposit date: | 1999-09-09 | | Release date: | 2000-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Sensory mechanism of oxygen sensor FixL from Rhizobium meliloti: crystallographic, mutagenesis and resonance Raman spectroscopic studies
J.MOL.BIOL., 301, 2000
|
|
5E8D
 
 | | Crystal structure of human epiregulin in complex with the Fab fragment of murine monoclonal antibody 9E5 | | Descriptor: | CHLORIDE ION, GLYCEROL, Proepiregulin, ... | | Authors: | Kado, Y, Mizohata, E, Nagatoishi, S, Iijima, M, Shinoda, K, Miyafusa, T, Nakayama, T, Yoshizumi, T, Sugiyama, A, Kawamura, T, Lee, Y.H, Matsumura, H, Doi, H, Fujitani, H, Kodama, T, Shibasaki, Y, Tsumoto, K, Inoue, T. | | Deposit date: | 2015-10-14 | | Release date: | 2015-12-09 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
J.Biol.Chem., 291, 2016
|
|
1MSE
 
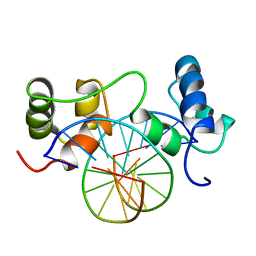 | | SOLUTION STRUCTURE OF A SPECIFIC DNA COMPLEX OF THE MYB DNA-BINDING DOMAIN WITH COOPERATIVE RECOGNITION HELICES | | Descriptor: | C-Myb DNA-Binding Domain, DNA (5'-D(*AP*TP*GP*TP*GP*TP*GP*TP*CP*AP*GP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*AP*AP*CP*TP*GP*AP*CP*AP*CP*AP*CP*AP*T)-3') | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Sekikawa, A, Inoue, T, Kanai, H, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-01-24 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices.
Cell(Cambridge,Mass.), 79, 1994
|
|
6L98
 
 | | Crystalline cast nephropathy-causing Bence-Jones protein AK: An entire immunoglobulin lambda light chain dimer | | Descriptor: | Bence-Jones protein lambda light chain AK | | Authors: | Nakagaki, T, Noguchi, K, Yohda, M, Odaka, M, Wakui, H, Matsumura, H. | | Deposit date: | 2019-11-08 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Multiple Myeloma-Associated Ig Light Chain Crystalline Cast Nephropathy.
Kidney Int Rep, 5, 2020
|
|
7Y8Q
 
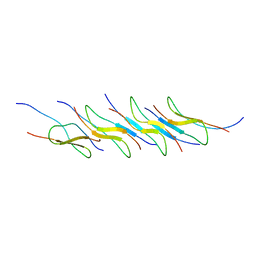 | | Amyloid-beta assemblage on GM1-containing membranes | | Descriptor: | Amyloid-beta protein 40 | | Authors: | Yagi-Utsumi, M, Itoh, S.G, Okumura, H, Yanagisawa, K, Kato, K, Nishimura, K. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The Double-Layered Structure of Amyloid-beta Assemblage on GM1-Containing Membranes Catalytically Promotes Fibrillization.
Acs Chem Neurosci, 14, 2023
|
|
7XZR
 
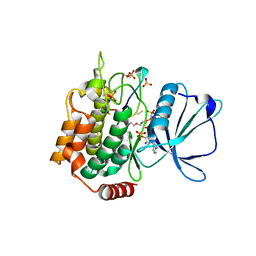 | | Crystal structure of TNIK-AMPPNP-thiopeptide TP15 complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, ... | | Authors: | Hamada, K, Vinogradov, A.A, Zhang, Y, Chang, J.S, Nishimura, H, Goto, Y, Onaka, H, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | De Novo Discovery of Thiopeptide Pseudo-natural Products Acting as Potent and Selective TNIK Kinase Inhibitors.
J.Am.Chem.Soc., 144, 2022
|
|
7XZQ
 
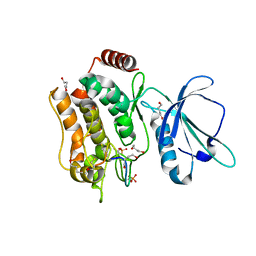 | | Crystal structure of TNIK-thiopeptide TP1 complex | | Descriptor: | 1,4-BUTANEDIOL, TRAF2 and NCK-interacting protein kinase, thiopeptide TP1 | | Authors: | Hamada, K, Vinogradov, A.A, Zhang, Y, Chang, J.S, Nishimura, H, Goto, Y, Onaka, H, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | De Novo Discovery of Thiopeptide Pseudo-natural Products Acting as Potent and Selective TNIK Kinase Inhibitors.
J.Am.Chem.Soc., 144, 2022
|
|
7YQA
 
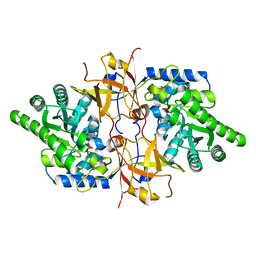 | | Crystal structure of D-threonine aldolase from Chlamydomonas reinhardtii | | Descriptor: | D-threonine aldolase, MAGNESIUM ION | | Authors: | Hirato, Y, Goto, M, Mizobuchi, T, Muramatsu, H, Tanigawa, M, Nishimura, K. | | Deposit date: | 2022-08-05 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of pyridoxal 5'-phosphate-bound D-threonine aldolase from Chlamydomonas reinhardtii.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
1RIL
 
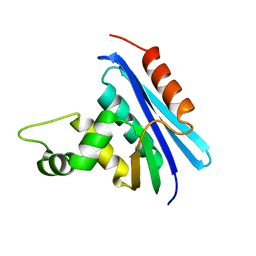 | | CRYSTAL STRUCTURE OF RIBONUCLEASE H FROM THERMUS THERMOPHILUS HB8 REFINED AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Okumura, M, Katayanagi, K, Kimura, S, Kanaya, S, Nakamura, H, Morikawa, K. | | Deposit date: | 1993-01-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ribonuclease H from Thermus thermophilus HB8 refined at 2.8 A resolution.
J.Mol.Biol., 230, 1993
|
|
1MSF
 
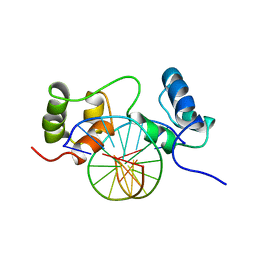 | | SOLUTION STRUCTURE OF A SPECIFIC DNA COMPLEX OF THE MYB DNA-BINDING DOMAIN WITH COOPERATIVE RECOGNITION HELICES | | Descriptor: | C-Myb DNA-Binding Domain, DNA (5'-D(*AP*TP*GP*TP*GP*TP*GP*TP*CP*AP*GP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*AP*AP*CP*TP*GP*AP*CP*AP*CP*AP*CP*AP*T)-3') | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Sekikawa, A, Inoue, T, Kanai, H, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-01-24 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices.
Cell(Cambridge,Mass.), 79, 1994
|
|
2NVL
 
 | | Crystal structure of archaeal peroxiredoxin, thioredoxin peroxidase from Aeropyrum pernix K1 (sulfonic acid form) | | Descriptor: | Probable peroxiredoxin | | Authors: | Nakamura, T, Yamamoto, T, Abe, M, Matsumura, H, Hagihara, Y, Goto, T, Yamaguchi, T, Inoue, T. | | Deposit date: | 2006-11-13 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1PRU
 
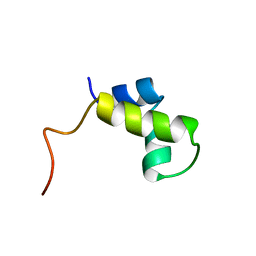 | | PURINE REPRESSOR DNA-BINDING DOMAIN DNA BINDING | | Descriptor: | PURINE REPRESSOR | | Authors: | Nagadoi, A, Morikawa, S, Nakamura, H, Enari, M, Kobayashi, K, Yamamoto, H, Sampei, G, Mizobuchi, K, Schumacher, M.A, Brennan, R.G, Nishimura, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural comparison of the free and DNA-bound forms of the purine repressor DNA-binding domain.
Structure, 3, 1995
|
|
1PRV
 
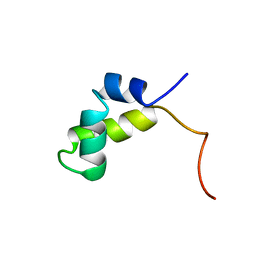 | | PURINE REPRESSOR DNA-BINDING DOMAIN DNA BINDING | | Descriptor: | PURINE REPRESSOR | | Authors: | Nagadoi, A, Morikawa, S, Nakamura, H, Enari, M, Kobayashi, K, Yamamoto, H, Sampei, G, Mizobuchi, K, Schumacher, M.A, Brennan, R.G, Nishimura, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural comparison of the free and DNA-bound forms of the purine repressor DNA-binding domain.
Structure, 3, 1995
|
|
