3KDP
 
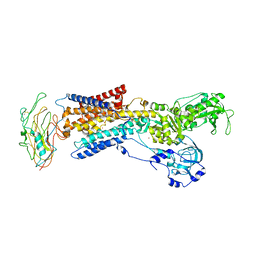 | | Crystal structure of the sodium-potassium pump | | Descriptor: | CHOLESTEROL, MAGNESIUM ION, Na+/K+ ATPase gamma subunit transcript variant a, ... | | Authors: | Morth, J.P, Pedersen, B.P, Nissen, P. | | Deposit date: | 2009-10-23 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the sodium-potassium pump.
Nature, 450, 2007
|
|
3B8E
 
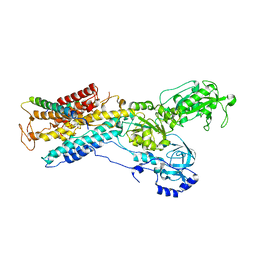 | | Crystal structure of the sodium-potassium pump | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, MAGNESIUM ION, Na+/K+ ATPase gamma subunit transcript variant a, ... | | Authors: | Morth, J.P, Pedersen, P.B, Toustrup-Jensen, M.S, Soerensen, T.L.M, Petersen, J, Andersen, J.P, Vilsen, B, Nissen, P. | | Deposit date: | 2007-11-01 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the sodium-potassium pump.
Nature, 450, 2007
|
|
6HCZ
 
 | |
1S8N
 
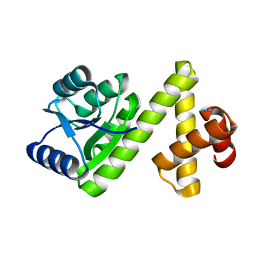 | | Crystal structure of Rv1626 from Mycobacterium tuberculosis | | Descriptor: | AZIDE ION, putative antiterminator | | Authors: | Morth, J.P, Feng, V, Perry, L.J, Svergun, D.I, Tucker, P.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-02-03 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | The Crystal and Solution Structure of a Putative Transcriptional Antiterminator from Mycobacterium tuberculosis.
Structure, 12, 2004
|
|
1SD5
 
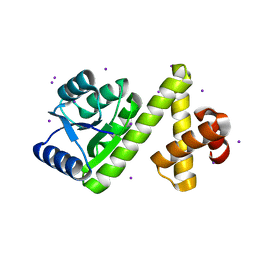 | | Crystal structure of Rv1626 | | Descriptor: | IODIDE ION, putative antiterminator | | Authors: | Morth, J.P, Feng, V, Perry, L.J, Svergun, D.I, Tucker, P.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-02-13 | | Release date: | 2004-09-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Crystal and Solution Structure of a Putative Transcriptional Antiterminator from Mycobacterium tuberculosis.
Structure, 12, 2004
|
|
8Q8H
 
 | |
8AMY
 
 | | High-resolution crystal structure of the Mu8.1 conotoxin from Conus Mucronatus | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BICARBONATE ION, ... | | Authors: | Mueller, E, Hackney, C.M, Ellgaard, L, Morth, J.P. | | Deposit date: | 2022-08-04 | | Release date: | 2023-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | High-resolution crystal structure of the Mu8.1 conotoxin from Conus mucronatus.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
5JHO
 
 | |
7Z6T
 
 | | Aspergillus clavatus M36 protease without the propeptide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Extracellular metalloproteinase mep, ... | | Authors: | Wilkens, C, Qiu, J, Meyer, A.S, Morth, J.P. | | Deposit date: | 2022-03-14 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Aspergillus clavatus M36 protease without the propeptide
To Be Published
|
|
5JJ2
 
 | | Crystal structure of the central domain of human AKAP18 gamma/delta in complex with malonate | | Descriptor: | A-kinase anchor protein 7 isoform gamma, MALONATE ION | | Authors: | Bjerregaard-Andersen, K, Ostensen, E, Scott, J.D, Tasken, K, Morth, J.P. | | Deposit date: | 2016-04-22 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Malonate in the nucleotide-binding site traps human AKAP18 gamma / delta in a novel conformational state.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5EZD
 
 | | Crystal structure of a Hepatocyte growth factor activator inhibitor-1 (HAI-1) fragment covering the PKD-like 'internal' domain and Kunitz domain 1 | | Descriptor: | ACETATE ION, Kunitz-type protease inhibitor 1, POTASSIUM ION | | Authors: | Hong, Z, Andreasen, P.A, Morth, J.P, Jensen, J.K. | | Deposit date: | 2015-11-26 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a Two-domain Fragment of Hepatocyte Growth Factor Activator Inhibitor-1: FUNCTIONAL INTERACTIONS BETWEEN THE KUNITZ-TYPE INHIBITOR DOMAIN-1 AND THE NEIGHBORING POLYCYSTIC KIDNEY DISEASE-LIKE DOMAIN.
J.Biol.Chem., 291, 2016
|
|
4HYF
 
 | | Structural basis and SAR for OD 270, a lead stage 1,2,4-triazole based specific Tankyrase1/2 inhibitor | | Descriptor: | 4-{5-[(E)-2-{4-(2-chlorophenyl)-5-[5-(methylsulfonyl)pyridin-2-yl]-4H-1,2,4-triazol-3-yl}ethenyl]-1,3,4-oxadiazol-2-yl}benzonitrile, NICOTINAMIDE, Tankyrase-2, ... | | Authors: | Perdreau, H, Ekblad, B, Voronkov, A, Holsworth, D.D, Waaler, J, Drewes, G, Schueler, H, Krauss, S, Morth, J.P. | | Deposit date: | 2012-11-13 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis and SAR for G007-LK, a Lead Stage 1,2,4-Triazole Based Specific Tankyrase 1/2 Inhibitor.
J.Med.Chem., 56, 2013
|
|
4H9G
 
 | | Probing EF-Tu with a very small brominated fragment library identifies the CCA pocket | | Descriptor: | 5-bromofuran-2-carboxylic acid, AMMONIUM ION, Elongation factor Tu-A, ... | | Authors: | Groftehauge, M.K, Therkelsen, M, Taaning, R.H, Skrydstrup, T, Morth, J.P, Nissen, P. | | Deposit date: | 2012-09-24 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Identifying ligand-binding hot spots in proteins using brominated fragments.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3JRY
 
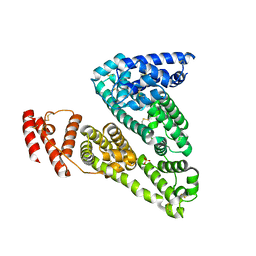 | | Human Serum albumin with bound Sulfate | | Descriptor: | SULFATE ION, Serum albumin | | Authors: | Hein, K.L, Kragh-Hansen, U, Morth, J.P, Nissen, P. | | Deposit date: | 2009-09-09 | | Release date: | 2010-04-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic analysis reveals a unique lidocaine binding site on human serum albumin.
J.Struct.Biol., 171, 2010
|
|
3JQZ
 
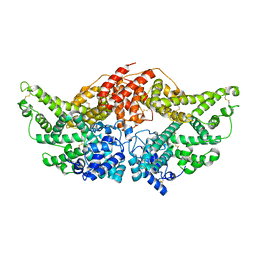 | | Crystal Structure of Human serum albumin complexed with Lidocaine | | Descriptor: | 2-(diethylamino)-N-(2,6-dimethylphenyl)ethanamide, Serum albumin | | Authors: | Hein, K.L, Kragh-Hansen, U, Morth, J.P, Nissen, P. | | Deposit date: | 2009-09-08 | | Release date: | 2010-04-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystallographic analysis reveals a unique lidocaine binding site on human serum albumin.
J.Struct.Biol., 2010
|
|
3BA6
 
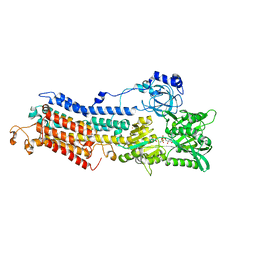 | | Structure of the Ca2E1P phosphoenzyme intermediate of the SERCA Ca2+-ATPase | | Descriptor: | AMP PHOSPHORAMIDATE, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Picard, M, Winther, A.M.L, Olesen, C, Gyrup, C, Morth, J.P, Oxvig, C, Moller, J.V, Nissen, P. | | Deposit date: | 2007-11-07 | | Release date: | 2007-12-18 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of calcium transport by the calcium pump.
Nature, 450, 2007
|
|
8BPD
 
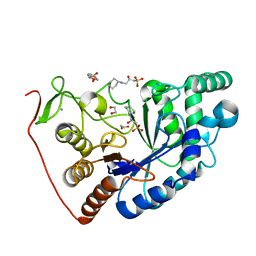 | | Structural and Functional Characterization of the Novel Endo-alpha(1,4)-Fucoidanase Mef1 from the Marine Bacterium Muricauda eckloniae | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Mikkelsen, M.D, Meyer, A.S, Morth, J.P. | | Deposit date: | 2022-11-16 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional characterization of the novel endo-alpha (1,4)-fucoidanase Mef1 from the marine bacterium Muricauda eckloniae.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
3Q02
 
 | |
5NNA
 
 | | Isatin hydrolase A (IHA) from Labrenzia aggregata bound to benzyl benzoate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZOIC ACID PHENYLMETHYLESTER, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sommer, T, Bjerregaard-Andersen, K, Morth, J.P. | | Deposit date: | 2017-04-08 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A fundamental catalytic difference between zinc and manganese dependent enzymes revealed in a bacterial isatin hydrolase.
Sci Rep, 8, 2018
|
|
3N23
 
 | | Crystal structure of the high affinity complex between ouabain and the E2P form of the sodium-potassium pump | | Descriptor: | MAGNESIUM ION, Na+/K+ ATPase gamma subunit transcript variant a, OUABAIN, ... | | Authors: | Yatime, L, Laursen, M, Morth, J.P, Esmann, M, Nissen, P, Fedosova, N.U. | | Deposit date: | 2010-05-17 | | Release date: | 2011-01-19 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Structural insights into the high affinity binding of cardiotonic steroids to the Na+,K+-ATPase.
J.Struct.Biol., 174, 2011
|
|
3N5K
 
 | | Structure Of The (Sr)Ca2+-ATPase E2-AlF4- Form | | Descriptor: | ACETATE ION, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Bublitz, M, Olesen, C, Poulsen, H, Morth, J.P, Moller, J.V, Nissen, P. | | Deposit date: | 2010-05-25 | | Release date: | 2011-06-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ion pathways in the sarcoplasmic reticulum Ca2+-ATPase.
J.Biol.Chem., 288, 2013
|
|
5NNB
 
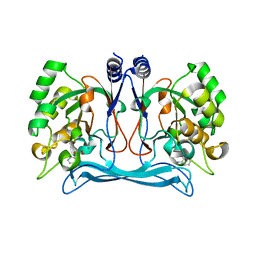 | |
3N8G
 
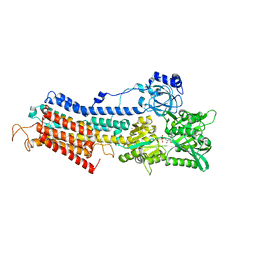 | | Structure of the (SR)Ca2+-ATPase Ca2-E1-CaAMPPCP form | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Bublitz, M, Olesen, C, Poulsen, H, Morth, J.P, Moller, J.V, Nissen, P. | | Deposit date: | 2010-05-28 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.585 Å) | | Cite: | Ion pathways in the sarcoplasmic reticulum Ca2+-ATPase.
J.Biol.Chem., 288, 2013
|
|
3RFU
 
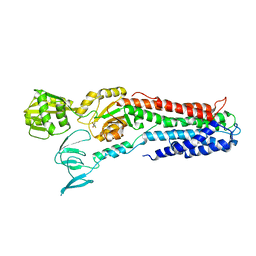 | | Crystal structure of a copper-transporting PIB-type ATPase | | Descriptor: | Copper efflux ATPase, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Gourdon, P, Liu, X, Skjorringe, T, Morth, J.P, Birk Moller, L, Panyella Pedersen, B, Nissen, P. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a copper-transporting PIB-type ATPase.
Nature, 475, 2011
|
|
5NMP
 
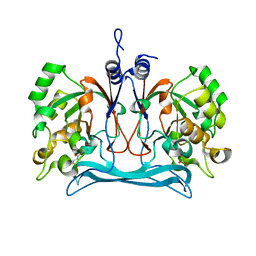 | |
