6ALK
 
 | | NMR solution structure of the major beech pollen allergen Fag s 1 | | Descriptor: | Fag s 1 pollen allergen | | Authors: | Moraes, A.H, Asam, A, Almeida, F.C.L, Wallner, M, Ferreira, F, Valente, A.P. | | Deposit date: | 2017-08-08 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for cross-reactivity and conformation fluctuation of the major beech pollen allergen Fag s 1.
Sci Rep, 8, 2018
|
|
2MBX
 
 | | Structure, dynamics and stability of allergen cod parvalbumin Gad m 1 by solution and high-pressure NMR. | | Descriptor: | CALCIUM ION, Parvalbumin beta | | Authors: | Moraes, A.H, Ackerbauer, D, Bublin, M, Ferreira, F, Almeida, F.C.L, Breiteneder, H, Valente, A. | | Deposit date: | 2013-08-07 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution and high-pressure NMR studies of the structure, dynamics, and stability of the cross-reactive allergenic cod parvalbumin Gad m 1.
Proteins, 82, 2014
|
|
1WVA
 
 | | Crystal structure of human arginase I from twinned crystal | | Descriptor: | Arginase 1, MANGANESE (II) ION, S-2-(BORONOETHYL)-L-CYSTEINE | | Authors: | Di Costanzo, L, Sabio, G, Mora, A, Rodriguez, P.C, Ochoa, A.C, Centeno, F, Christianson, D.W. | | Deposit date: | 2004-12-14 | | Release date: | 2005-09-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of human arginase I at 1.29 A resolution and exploration of inhibition in the immune response
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AEB
 
 | | Crystal structure of human arginase I at 1.29 A resolution and exploration of inhibition in immune response. | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, Arginase 1, MANGANESE (II) ION | | Authors: | Di Costanzo, L, Sabio, G, Mora, A, Rodriguez, P.C, Ochoa, A.C, Centeno, F, Christianson, D.W. | | Deposit date: | 2005-07-21 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystal structure of human arginase I at 1.29 A resolution and exploration of inhibition in the immune response.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1WVB
 
 | | Crystal structure of human arginase I: the mutant E256Q | | Descriptor: | Arginase 1, MANGANESE (II) ION, S-2-(BORONOETHYL)-L-CYSTEINE | | Authors: | Di Costanzo, L, Guadalupe, S, Mora, A, Centeno, F, Christianson, D.W. | | Deposit date: | 2004-12-14 | | Release date: | 2005-09-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human arginase I: the mutant E256Q
To be Published
|
|
2N5A
 
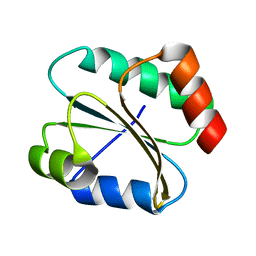 | |
2N5B
 
 | |
6XIO
 
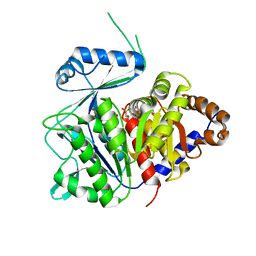 | | ADP-dependent kinase complex with fructose-6-phosphate and ADPbetaS | | Descriptor: | 5'-O-[(R)-HYDROXY(THIOPHOSPHONOOXY)PHOSPHORYL]ADENOSINE, 6-O-phosphono-beta-D-fructofuranose, ADP-dependent phosphofructokinase, ... | | Authors: | Munoz, S, Gonzalez-Ordenes, F, Fuentes, N, Maturana, P, Herrera-Morande, A, Villalobos, P, Castro-Fernandez, V. | | Deposit date: | 2020-06-20 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structure of an ancestral ADP-dependent kinase with fructose-6P reveals key residues for binding, catalysis, and ligand-induced conformational changes.
J.Biol.Chem., 296, 2020
|
|
6XAT
 
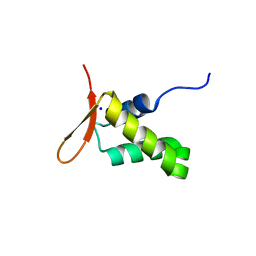 | | Crystal structure of the human FoxP4 DNA binding Domain | | Descriptor: | FOXP4 protein, SODIUM ION | | Authors: | VIllalobos, P, Castro-Fernandez, V, Medina, E, Gonzalez-Ordenes, F, Maturana, P, Herrera-Morande, A, Ramirez-Sarmiento, C.A, Babul, J. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unraveling the folding and dimerization properties of the human FoxP subfamily of transcription factors.
Febs Lett., 597, 2023
|
|
5KKG
 
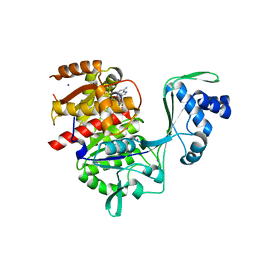 | | Crystal structure of E72A mutant of ancestral protein ancMT of ADP-dependent sugar kinases family | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, IODIDE ION, ... | | Authors: | Castro-Fernandez, V, Herrera-Morande, A, Zamora, R, Merino, F, Pereira, H.M, Brandao-Neto, J, Garratt, R, Guixe, V. | | Deposit date: | 2016-06-21 | | Release date: | 2017-07-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | Reconstructed ancestral enzymes reveal that negative selection drove the evolution of substrate specificity in ADP-dependent kinases.
J. Biol. Chem., 292, 2017
|
|
5K27
 
 | | Crystal structure of ancestral protein ancMT of ADP-dependent sugar kinases family. | | Descriptor: | ADENOSINE MONOPHOSPHATE, IODIDE ION, ancMT | | Authors: | Castro-Fernandez, V, Herrera-Morande, A, Zamora, R, Merino, F, Pereira, H.M, Brandao-Neto, J, Garratt, R, Guixe, V. | | Deposit date: | 2016-05-18 | | Release date: | 2017-05-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Reconstructed ancestral enzymes reveal that negative selection drove the evolution of substrate specificity in ADP-dependent kinases.
J. Biol. Chem., 292, 2017
|
|
7P35
 
 | | Structure of the SARS-CoV-2 3CL protease in complex with rupintrivir | | Descriptor: | 3C-like proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Fabrega-Ferrer, M, Perez-Saavedra, J, Herrera-Morande, A, Coll, M. | | Deposit date: | 2021-07-07 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.256 Å) | | Cite: | Structure and inhibition of SARS-CoV-1 and SARS-CoV-2 main proteases by oral antiviral compound AG7404.
Antiviral Res., 208, 2022
|
|
4IYD
 
 | |
4IYF
 
 | |
7ZQV
 
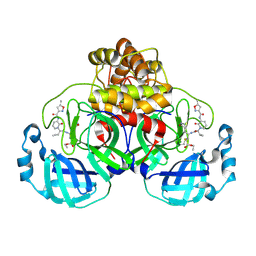 | | Structure of the SARS-CoV-2 main protease in complex with AG7404 | | Descriptor: | 3C-like proteinase nsp5, ethyl (4R)-4-({(2S)-2-[3-{[(5-methyl-1,2-oxazol-3-yl)carbonyl]amino}-2-oxopyridin-1(2H)-yl]pent-4-ynoyl}amino)-5-[(3S)-2-oxopyrrolidin-3-yl]pentanoate | | Authors: | Fabrega-Ferrer, M, Herrera-Morande, A, Perez-Saavedra, J, Coll, M. | | Deposit date: | 2022-05-03 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure and inhibition of SARS-CoV-1 and SARS-CoV-2 main proteases by oral antiviral compound AG7404.
Antiviral Res., 208, 2022
|
|
7ZQW
 
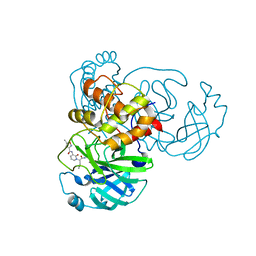 | | Structure of the SARS-CoV-1 main protease in complex with AG7404 | | Descriptor: | 3C-like proteinase nsp5, ethyl (4R)-4-({(2S)-2-[3-{[(5-methyl-1,2-oxazol-3-yl)carbonyl]amino}-2-oxopyridin-1(2H)-yl]pent-4-ynoyl}amino)-5-[(3S)-2-oxopyrrolidin-3-yl]pentanoate | | Authors: | Muriel-Goni, S, Fabrega-Ferrer, M, Herrera-Morande, A, Coll, M. | | Deposit date: | 2022-05-03 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure and inhibition of SARS-CoV-1 and SARS-CoV-2 main proteases by oral antiviral compound AG7404.
Antiviral Res., 208, 2022
|
|
6XEQ
 
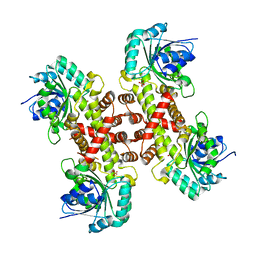 | | Crystal structure of the tetrameric 6-phosphogluconate dehydrogenase from Gluconobacter oxidans | | Descriptor: | 6-phosphogluconate dehydrogenase, SULFATE ION | | Authors: | Maturana, P, Roversi, P, Castro-Fernandez, V, Herrera-Morande, A, Garratt, R.C, Cabrera, R. | | Deposit date: | 2020-06-13 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the 6-phosphogluconate dehydrogenase from Gluconobacter oxydans reveals tetrameric 6PGDHs as the crucial intermediate in the evolution of structure and cofactor preference in the 6PGDH family [version 1; peer review: 1 approved, 1 approved with reservations]
Wellcome Open Res, 6, 2021
|
|
5O5X
 
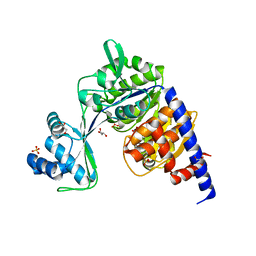 | | Crystal structure of Thermococcus litoralis ADP-dependent glucokinase (GK) | | Descriptor: | ADP-dependent glucokinase,ADP-dependent glucokinase,ADP-dependent glucokinase, GLYCEROL, SULFATE ION | | Authors: | Herrera-Morande, A, Castro-Fernandez, V, Merino, F, Ramirez-Sarmiento, C.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2017-06-02 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Protein topology determines substrate-binding mechanism in homologous enzymes.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5O5Z
 
 | | CRYSTAL STRUCTURE OF THERMOCOCCUS LITORALIS ADP-DEPENDENT GLUCOKINASE (GK) | | Descriptor: | 5'-O-[(R)-HYDROXY(THIOPHOSPHONOOXY)PHOSPHORYL]ADENOSINE, ADP-dependent glucokinase,ADP-dependent glucokinase,ADP-dependent glucokinase, GLYCEROL, ... | | Authors: | Herrera-Morande, A, Castro-Fernandez, V, Merino, F, Ramirez-Sarmiento, C.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2017-06-02 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.441 Å) | | Cite: | Protein topology determines substrate-binding mechanism in homologous enzymes.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
4B8R
 
 | | Crystal Structure of Thermococcus litoralis ADP-dependent Glucokinase (GK) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADP-DEPENDENT GLUCOKINASE, ... | | Authors: | Herrera-Morande, A, Rivas-Pardo, J.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2012-08-30 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure, Saxs and Kinetic Mechanism of Hyperthermophilic Adp-Dependent Glucokinase from Thermococcus Litoralis Reveal a Conserved Mechanism for Catalysis.
Plos One, 8, 2013
|
|
5O5Y
 
 | | Crystal structure of Thermococcus litoralis ADP-dependent glucokinase (GK) | | Descriptor: | ADP-dependent glucokinase,ADP-dependent glucokinase,ADP-dependent glucokinase, GLYCEROL, TRIETHYLENE GLYCOL, ... | | Authors: | Herrera-Morande, A, Castro-Fernandez, V, Merino, F, Ramirez-Sarmiento, C.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2017-06-02 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.915 Å) | | Cite: | Protein topology determines substrate-binding mechanism in homologous enzymes.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
7L07
 
 | | Last common ancestor of HMPPK and PLK/HMPPK vitamin kinases | | Descriptor: | ALUMINUM FLUORIDE, Ancestral Protein AncC | | Authors: | Gonzalez-Ordenes, F, Maturana, P, Herrera-Morande, A, Araya, G, Arizabalos, S, Castro-Fernandez, V. | | Deposit date: | 2020-12-11 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and molecular dynamics simulations of a promiscuous ancestor reveal residues and an epistatic interaction involved in substrate binding and catalysis in the ATP-dependent vitamin kinase family members.
Protein Sci., 30, 2021
|
|
6C8Z
 
 | | Last common ancestor of ADP-dependent phosphofructokinases from Methanosarcinales | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADP-dependent phosphofructokinase, MAGNESIUM ION, ... | | Authors: | Castro-Fernandez, V, Gonzalez-Ordenes, F, Munoz, S, Fuentes, N, Leonardo, D, Fuentealba, M, Herrera-Morande, A, Maturana, P, Villalobos, P, Garratt, R. | | Deposit date: | 2018-01-25 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | ADP-Dependent Kinases From the Archaeal OrderMethanosarcinalesAdapt to Salt by a Non-canonical Evolutionarily Conserved Strategy.
Front Microbiol, 9, 2018
|
|
4B8S
 
 | | Crystal Structure of Thermococcus litoralis ADP-dependent Glucokinase (GK) | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP-DEPENDENT GLUCOKINASE, GLYCEROL, ... | | Authors: | Herrera-Morande, A, Rivas-Pardo, J.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2012-08-30 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal Structure, Saxs and Kinetic Mechanism of Hyperthermophilic Adp-Dependent Glucokinase from Thermococcus Litoralis Reveal a Conserved Mechanism for Catalysis.
Plos One, 8, 2013
|
|
4JE9
 
 | | Crystal structure of an engineered metal-free RIDC1 construct with four interfacial disulfide bonds | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Soluble cytochrome b562 | | Authors: | Tezcan, F.A, Medina-Morales, A.M, Perez, A, Brodin, J.D. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | In Vitro and Cellular Self-Assembly of a Zn-Binding Protein Cryptand via Templated Disulfide Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
