8DNK
 
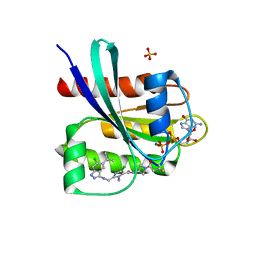 | | Crystal structure of human KRAS G12C covalently bound with Taiho WO2020/085493A1 compound 6 | | Descriptor: | 2-{[(5-tert-butyl-6-chloro-1H-indazol-3-yl)amino]methyl}-4-chloro-1-methyl-N-(1-propanoylazetidin-3-yl)-1H-imidazole-5-carboxamide, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Mohr, C. | | Deposit date: | 2022-07-11 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Modeling receptor flexibility in the structure-based design of KRAS G12C inhibitors.
J.Comput.Aided Mol.Des., 36, 2022
|
|
8DNI
 
 | | Crystal structure of human KRAS G12C covalently bound with Araxes WO2020/028706A1 compound I-1 | | Descriptor: | (4P)-4-(5-methyl-1H-indazol-4-yl)-6-(2-propanoyl-2,6-diazaspiro[3.4]octan-6-yl)-2-(pyrrolidin-1-yl)pyrimidine-5-carbonitrile, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mohr, C. | | Deposit date: | 2022-07-11 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Modeling receptor flexibility in the structure-based design of KRAS G12C inhibitors.
J.Comput.Aided Mol.Des., 36, 2022
|
|
8DNJ
 
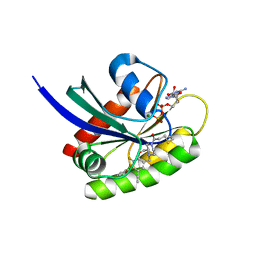 | | Crystal structure of human KRAS G12C covalently bound with AstraZeneca WO2020/178282A1 compound 76 | | Descriptor: | 1-[(5S,9P,12aR)-9-(2-chloro-6-hydroxyphenyl)-8-ethynyl-10-fluoro-3,4,12,12a-tetrahydro-6H-pyrazino[2,1-c][1,4]benzoxazepin-2(1H)-yl]propan-1-one, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Mohr, C. | | Deposit date: | 2022-07-11 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Modeling receptor flexibility in the structure-based design of KRAS G12C inhibitors.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5IPJ
 
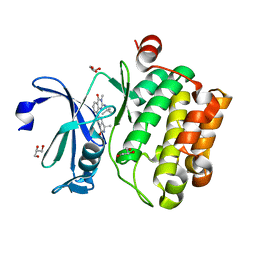 | | Crystal structure of human Pim-1 kinase in complex with a quinazolinone-pyrrolopyrrolone inhibitor. | | Descriptor: | 2-(tert-butylamino)-3-methyl-8-[(6R)-6-methyl-4-oxo-1,4,5,6-tetrahydropyrrolo[3,4-b]pyrrol-2-yl]quinazolin-4(3H)-one, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Mohr, C. | | Deposit date: | 2016-03-09 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Optimization of Quinazolinone-pyrrolopyrrolones as Potent and Orally Bioavailable Pan-Pim Kinase Inhibitors.
J.Med.Chem., 59, 2016
|
|
3LHJ
 
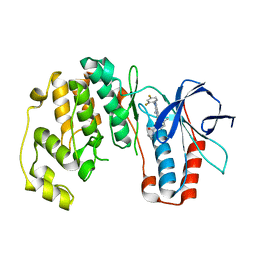 | | Crystal Structure of p38a Mitogen-Activated Protein Kinase in Complex with a Pyrazolopyridinone Inhibitor. | | Descriptor: | Mitogen-activated protein kinase 14, N-cyclopropyl-3-[1-(2,4-difluorophenyl)-7-methyl-6-oxo-6,7-dihydro-1H-pyrazolo[3,4-b]pyridin-5-yl]-4-methylbenzamide | | Authors: | Mohr, C, Jordan, S. | | Deposit date: | 2010-01-22 | | Release date: | 2010-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Discovery and evaluation of 7-alkyl-1,5-bis-aryl-pyrazolopyridinones as highly potent, selective, and orally efficacious inhibitors of p38alpha mitogen-activated protein kinase.
J.Med.Chem., 53, 2010
|
|
5KZI
 
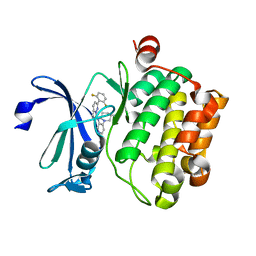 | | Crystal structure of human Pim-1 kinase in complex with an imidazopyridazine inhibitor. | | Descriptor: | Serine/threonine-protein kinase pim-1, ~{N}-[4-[(3~{S})-3-azanylpiperidin-1-yl]pyridin-3-yl]-2-[2,6-bis(fluoranyl)phenyl]imidazo[1,5-b]pyridazin-7-amine | | Authors: | Mohr, C. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of imidazopyridazines as potent Pim-1/2 kinase inhibitors.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
5EOL
 
 | |
6MT0
 
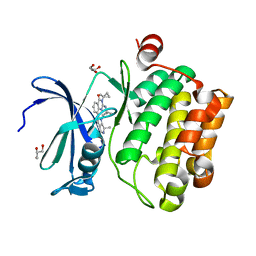 | | Crystal structure of human Pim-1 kinase in complex with a quinazolinone-pyrrolodihydropyrrolone inhibitor | | Descriptor: | 3-(1-methylcyclopropyl)-2-[(1-methylcyclopropyl)amino]-8-[(6R)-6-methyl-4-oxo-1,4,5,6-tetrahydropyrrolo[3,4-b]pyrrol-2-yl]quinazolin-4(3H)-one, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Mohr, C. | | Deposit date: | 2018-10-18 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of ( R)-8-(6-Methyl-4-oxo-1,4,5,6-tetrahydropyrrolo[3,4- b]pyrrol-2-yl)-3-(1-methylcyclopropyl)-2-((1-methylcyclopropyl)amino)quinazolin-4(3 H)-one, a Potent and Selective Pim-1/2 Kinase Inhibitor for Hematological Malignancies.
J. Med. Chem., 62, 2019
|
|
6OIM
 
 | |
4WSY
 
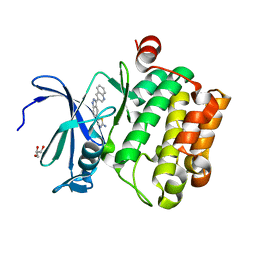 | |
4WT6
 
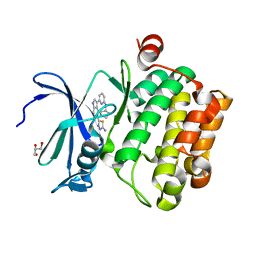 | |
4WRS
 
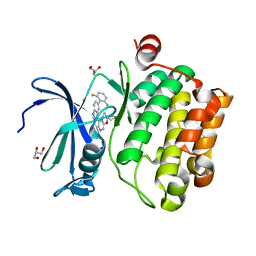 | |
4U6R
 
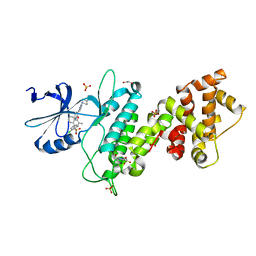 | | Crystal structure of human IRE1 cytoplasmic domains in complex with a sulfonamide inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-{4-[(3-{2-[(trans-4-aminocyclohexyl)amino]pyrimidin-4-yl}pyridin-2-yl)oxy]-3-methylnaphthalen-1-yl}-2-chlorobenzenesulfonamide, ... | | Authors: | Mohr, C. | | Deposit date: | 2014-07-29 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unfolded Protein Response in Cancer: IRE1 alpha Inhibition by Selective Kinase Ligands Does Not Impair Tumor Cell Viability.
Acs Med.Chem.Lett., 6, 2015
|
|
4U79
 
 | | Crystal structure of human JNK3 in complex with a benzenesulfonamide inhibitor. | | Descriptor: | Mitogen-activated protein kinase 10, N-{4-[(3-{2-[(trans-4-aminocyclohexyl)amino]pyrimidin-4-yl}pyridin-2-yl)oxy]naphthalen-1-yl}benzenesulfonamide | | Authors: | Mohr, C. | | Deposit date: | 2014-07-30 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Unfolded Protein Response in Cancer: IRE1 alpha Inhibition by Selective Kinase Ligands Does Not Impair Tumor Cell Viability.
Acs Med.Chem.Lett., 6, 2015
|
|
4TY1
 
 | |
3U8W
 
 | | Crystal Structure of p38a Mitogen-Activated Protein Kinase in Complex with a Triazolopyridazinone inhibitor | | Descriptor: | 3-[3-(2-chloro-6-fluorophenyl)-5-ethyl-6-oxo-5,6-dihydro[1,2,4]triazolo[4,3-b]pyridazin-7-yl]-N-cyclopropyl-4-methylbenzamide, Mitogen-activated protein kinase 14 | | Authors: | Mohr, C, Jordan, S. | | Deposit date: | 2011-10-17 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of triazolopyridazinones as potent p38alpha inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6UMI
 
 | | Crystal structure of erenumab Fab-b | | Descriptor: | erenumab Fab heavy chain, IgG1, erenumab Fab light chain | | Authors: | Mohr, C. | | Deposit date: | 2019-10-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Insight into Recognition of the CGRPR Complex by Migraine Prevention Therapy Aimovig (Erenumab).
Cell Rep, 30, 2020
|
|
6P8Z
 
 | | Crystal structure of human KRAS G12C covalently bound to an acryloylazetidine acetamide inhibitor | | Descriptor: | 2-[5-chloro-2-cyclopropyl-3-(5-methoxy-3,4-dihydroisoquinoline-2(1H)-carbonyl)-7-methyl-1H-indol-1-yl]-N-(1-propanoylazetidin-3-yl)acetamide, CALCIUM ION, GTPase KRas, ... | | Authors: | Mohr, C. | | Deposit date: | 2019-06-08 | | Release date: | 2019-08-28 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery ofN-(1-Acryloylazetidin-3-yl)-2-(1H-indol-1-yl)acetamides as Covalent Inhibitors of KRASG12C.
Acs Med.Chem.Lett., 10, 2019
|
|
6UMH
 
 | | Crystal structure of erenumab Fab-a | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3-PROPANDIOL, PHOSPHATE ION, ... | | Authors: | Mohr, C. | | Deposit date: | 2019-10-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular Insight into Recognition of the CGRPR Complex by Migraine Prevention Therapy Aimovig (Erenumab).
Cell Rep, 30, 2020
|
|
6UMJ
 
 | | Crystal structure of erenumab Fab-c | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3-BUTANEDIOL, erenumab Fab heavy chain, ... | | Authors: | Mohr, C. | | Deposit date: | 2019-10-09 | | Release date: | 2020-02-12 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Insight into Recognition of the CGRPR Complex by Migraine Prevention Therapy Aimovig (Erenumab).
Cell Rep, 30, 2020
|
|
6UMG
 
 | | Crystal structure of erenumab Fab bound to the extracellular domain of CGRP receptor | | Descriptor: | Calcitonin gene-related peptide type 1 receptor, Receptor activity-modifying protein 1, erenumab Fab heavy chain, ... | | Authors: | Mohr, C. | | Deposit date: | 2019-10-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Insight into Recognition of the CGRPR Complex by Migraine Prevention Therapy Aimovig (Erenumab).
Cell Rep, 30, 2020
|
|
6P8W
 
 | |
6P8Y
 
 | |
6P8X
 
 | |
6PGO
 
 | | Crystal structure of human KRAS G12C covalently bound to a phthalazine inhibitor | | Descriptor: | 1-{4-[7-chloro-6-(2-fluoro-6-hydroxyphenyl)-4-phenylphthalazin-1-yl]piperazin-1-yl}propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mohr, C. | | Deposit date: | 2019-06-24 | | Release date: | 2019-12-25 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of a Covalent Inhibitor of KRASG12C(AMG 510) for the Treatment of Solid Tumors.
J.Med.Chem., 63, 2020
|
|
