5LI1
 
 | | Structure of a Par3-inhibitory peptide bound to PKCiota core kinase domain | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Soriano, E.V, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | aPKC Inhibition by Par3 CR3 Flanking Regions Controls Substrate Access and Underpins Apical-Junctional Polarization.
Dev.Cell, 38, 2016
|
|
1BDY
 
 | | C2 DOMAIN FROM PROTEIN KINASE C DELTA | | Descriptor: | PROTEIN KINASE C | | Authors: | Pappa, H, Murray-Rust, J, Dekker, L.V, Parker, P.J, Mcdonald, N.Q. | | Deposit date: | 1998-05-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the C2 domain from protein kinase C-delta.
Structure, 6, 1998
|
|
1BUN
 
 | | STRUCTURE OF BETA2-BUNGAROTOXIN: POTASSIUM CHANNEL BINDING BY KUNITZ MODULES AND TARGETED PHOSPHOLIPASE ACTION | | Descriptor: | BETA2-BUNGAROTOXIN, SODIUM ION | | Authors: | Kwong, P.D, Mcdonald, N.Q, Sigler, P.B, Hendrickson, W.A. | | Deposit date: | 1995-10-15 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of beta 2-bungarotoxin: potassium channel binding by Kunitz modules and targeted phospholipase action.
Structure, 3, 1995
|
|
2BHN
 
 | | XPF from Aeropyrum pernix | | Descriptor: | XPF ENDONUCLEASE | | Authors: | Newman, M, Murray-Rust, J, Lally, J, Rudolf, J, Fadden, A, Knowles, P.P, White, M.F, McDonald, N.Q. | | Deposit date: | 2005-01-14 | | Release date: | 2005-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition.
EMBO J., 24, 2005
|
|
2BGW
 
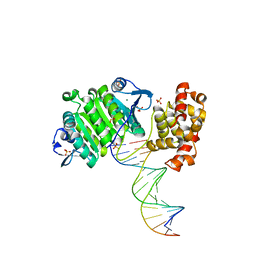 | | XPF from Aeropyrum pernix, complex with DNA | | Descriptor: | 5'-D(*GP*AP*TP*CP*AP*CP*AP*GP*AP*TP *GP*CP*TP*GP*A)-3', 5'-D(*TP*CP*AP*GP*CP*AP*TP*CP*TP*GP *TP*GP*AP*TP*C)-3', MAGNESIUM ION, ... | | Authors: | Newman, M, Murray-Rust, J, Lally, J, Rudolf, J, Fadden, A, Knowles, P.P, White, M.F, McDonald, N.Q. | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition.
EMBO J., 24, 2005
|
|
1YZB
 
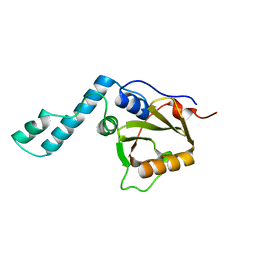 | | Solution structure of the Josephin domain of Ataxin-3 | | Descriptor: | Machado-Joseph disease protein 1 | | Authors: | Nicastro, G, Masino, L, Menon, R.P, Knowles, P.P, McDonald, N.Q, Pastore, A. | | Deposit date: | 2005-02-28 | | Release date: | 2005-07-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Josephin domain of ataxin-3: Structural determinants for molecular recognition
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2IVS
 
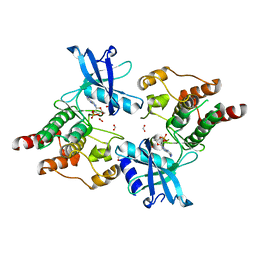 | | Crystal structure of non-phosphorylated RET tyrosine kinase domain | | Descriptor: | 2',3'- cyclic AMP, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Knowles, P.P, Murray-Rust, J, McDonald, N.Q. | | Deposit date: | 2006-06-16 | | Release date: | 2006-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Chemical Inhibition of the Ret Tyrosine Kinase Domain.
J.Biol.Chem., 281, 2006
|
|
2IVT
 
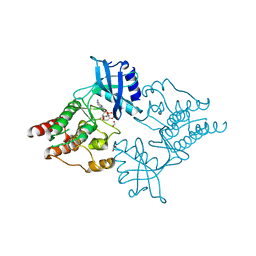 | |
2IVV
 
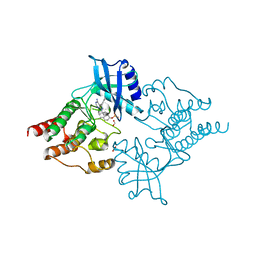 | | Crystal structure of phosphorylated RET tyrosine kinase domain complexed with the inhibitor PP1 | | Descriptor: | 1-TER-BUTYL-3-P-TOLYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET PRECURSOR | | Authors: | Knowles, P.P, Murray-Rust, J, McDonald, N.Q. | | Deposit date: | 2006-06-16 | | Release date: | 2006-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and chemical inhibition of the RET tyrosine kinase domain.
J. Biol. Chem., 281, 2006
|
|
2IVU
 
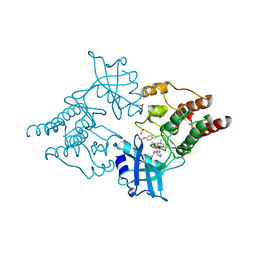 | |
2IYB
 
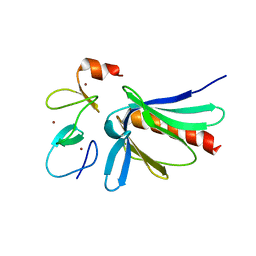 | |
2J16
 
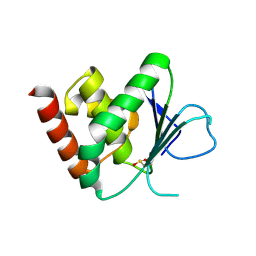 | | Apo & Sulphate bound forms of SDP-1 | | Descriptor: | MAGNESIUM ION, SULFATE ION, TYROSINE-PROTEIN PHOSPHATASE YIL113W | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2006-08-09 | | Release date: | 2007-05-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Redox-mediated substrate recognition by Sdp1 defines a new group of tyrosine phosphatases.
Nature, 447, 2007
|
|
2MC3
 
 | |
2J17
 
 | | pTyr bound form of SDP-1 | | Descriptor: | MAGNESIUM ION, O-PHOSPHOTYROSINE, TYROSINE-PROTEIN PHOSPHATASE YIL113W | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2006-08-09 | | Release date: | 2007-05-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Redox-mediated substrate recognition by Sdp1 defines a new group of tyrosine phosphatases.
Nature, 447, 2007
|
|
2V52
 
 | | Structure of MAL-RPEL2 complexed to G-actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Guettler, S, Langer, C.A, Treisman, R, McDonald, N.Q. | | Deposit date: | 2008-10-01 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular basis for G-actin binding to RPEL motifs from the serum response factor coactivator MAL.
EMBO J., 27, 2008
|
|
2JAJ
 
 | | DDAH1 complexed with L-257 | | Descriptor: | NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1, N~5~-{IMINO[(2-METHOXYETHYL)AMINO]METHYL}-L-ORNITHINE | | Authors: | Murray-Rust, J, O'Hara, B.P, Rossiter, S, Leiper, J.M, Vallance, P, McDonald, N.Q. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disruption of methylarginine metabolism impairs vascular homeostasis.
Nat. Med., 13, 2007
|
|
2JAI
 
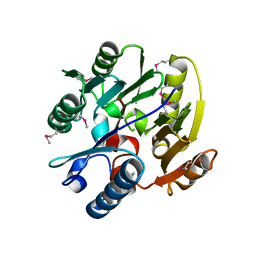 | | DDAH1 complexed with citrulline | | Descriptor: | CITRULLINE, NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1 | | Authors: | Murray-Rust, J, O'Hara, B.P, Rossiter, S, Leiper, J.M, Vallance, P, McDonald, N.Q. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Disruption of methylarginine metabolism impairs vascular homeostasis.
Nat. Med., 13, 2007
|
|
2V51
 
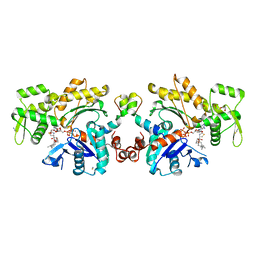 | | Structure of MAL-RPEL1 complexed to actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Guettler, S, Langer, C.A, Treisman, R, McDonald, N.Q. | | Deposit date: | 2008-10-01 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis for G-actin binding to RPEL motifs from the serum response factor coactivator MAL.
EMBO J., 27, 2008
|
|
2VT8
 
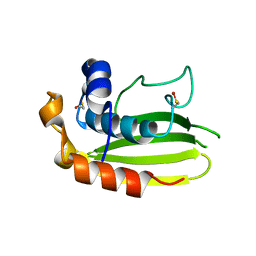 | | Structure of a conserved dimerisation domain within Fbox7 and PI31 | | Descriptor: | PROTEASOME INHIBITOR PI31 SUBUNIT | | Authors: | Kirk, R.J, Murray-Rust, J, Knowles, P.P, Laman, H, McDonald, N.Q. | | Deposit date: | 2008-05-12 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a Conserved Dimerization Domain within the F-Box Protein Fbxo7 and the Pi31 Proteasome Inhibitor.
J.Biol.Chem., 283, 2008
|
|
2YJF
 
 | | Oligomeric assembly of actin bound to MRTF-A | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Langer, C.A, Guettler, S, McDonald, N.Q, Treisman, R. | | Deposit date: | 2011-05-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a pentavalent G-actin*MRTF-A complex reveals how G-actin controls nucleocytoplasmic shuttling of a transcriptional coactivator.
Sci Signal, 4, 2011
|
|
2WH0
 
 | | Recognition of an intrachain tandem 14-3-3 binding site within protein kinase C epsilon | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, CALCIUM ION, PROTEIN KINASE C EPSILON TYPE, ... | | Authors: | Kostelecky, B, Saurin, A.T, Purkiss, A, Parker, P.J, McDonald, N.Q. | | Deposit date: | 2009-04-28 | | Release date: | 2009-08-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Recognition of an Intra-Chain Tandem 14-3-3 Binding Site within Pkc Epsilon.
Embo Rep., 10, 2009
|
|
4B1Y
 
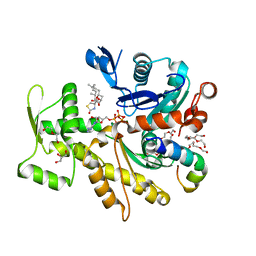 | | Structure of the Phactr1 RPEL-3 bound to G-actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
4B1X
 
 | | Structure of the Phactr1 RPEL-2 bound to G-actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
4B1U
 
 | | Structure of the Phactr1 RPEL domain and RPEL motif directed assemblies with G-actin reveal the molecular basis for actin binding cooperativity. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACTIN, ALPHA SKELETAL MUSCLE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
2X2L
 
 | | Crystal Structure of phosphorylated RET tyrosine kinase domain with inhibitor | | Descriptor: | (3Z)-5-AMINO-3-[(4-METHOXYPHENYL)METHYLIDENE]-1,3-DIHYDRO-2H-INDOL-2-ONE, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Knowles, P.P, Murray-Rust, J, Kjaer, S, McDonald, N.Q. | | Deposit date: | 2010-01-13 | | Release date: | 2010-02-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis, structure-activity relationship and crystallographic studies of 3-substituted indolin-2-one RET inhibitors.
Bioorg. Med. Chem., 18, 2010
|
|
