5GMY
 
 | |
2E5E
 
 | | Solution Structure of Variable-type Domain of Human Receptor for Advanced Glycation Endproducts | | Descriptor: | Advanced glycosylation end product-specific receptor | | Authors: | Matsumoto, S, Yoshida, T, Yasumatsu, I, Yamamoto, H, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2006-12-20 | | Release date: | 2007-12-25 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Variable-type Domain of Human Receptor for Advanced Glycation Endproducts
to be published
|
|
6R91
 
 | | Cryo-EM structure of NCP_THF2(-3)-UV-DDB | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R8Y
 
 | | Cryo-EM structure of NCP-6-4PP(-1)-UV-DDB | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R93
 
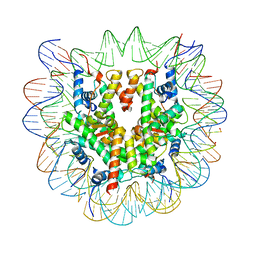 | | Cryo-EM structure of NCP-6-4PP | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-J, Histone H3.1, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R92
 
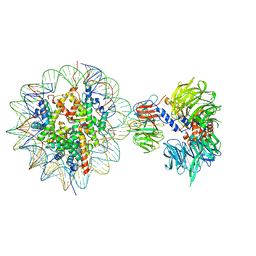 | | Cryo-EM structure of NCP-THF2(+1)-UV-DDB class B | | Descriptor: | DNA damage-binding protein 1,DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R8Z
 
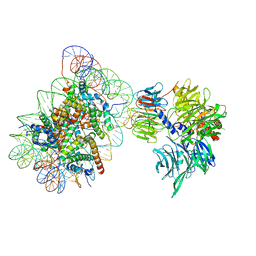 | | Cryo-EM structure of NCP_THF2(-1)-UV-DDB | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R94
 
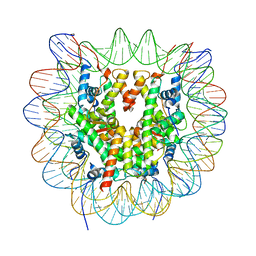 | | Cryo-EM structure of NCP_THF2(-3) | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-J, Histone H3.1, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R90
 
 | | Cryo-EM structure of NCP-THF2(+1)-UV-DDB class A | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
5ZC6
 
 | | Solution structure of H-RasT35S mutant protein in complex with KBFM123 | | Descriptor: | 3-oxidanyl-~{N}-[[(2~{R})-oxolan-2-yl]methyl]naphthalene-2-carboxamide, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Matsumoto, S, Hayashi, Y, Hiraga, T, Matsuo, K, Kataoka, T. | | Deposit date: | 2018-02-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis for Allosteric Inhibition of GTP-Bound H-Ras Protein by a Small-Molecule Compound Carrying a Naphthalene Ring
Biochemistry, 57, 2018
|
|
5X9S
 
 | | Crystal structure of fully modified H-Ras-GppNHp | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Matsumoto, S, Ke, H, Murashima, Y, Taniguchi-Tamura, H, Miyamoto, R, Yoshikawa, Y, Kumasaka, T, Mizohata, E, Edamatsu, H, Kataoka, T. | | Deposit date: | 2017-03-09 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for intramolecular interaction of post-translationally modified H-RasGTP prepared by protein ligation
FEBS Lett., 591, 2017
|
|
3VGP
 
 | | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (AF_0329) from Archaeoglobus fulgidus | | Descriptor: | Transmembrane oligosaccharyl transferase, putative | | Authors: | Matsumoto, S, Igura, M, Nyirenda, J, Yuzawa, S, Noda, N.N, Inagaki, F, Kohda, D. | | Deposit date: | 2011-08-18 | | Release date: | 2012-07-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the C-Terminal Globular Domain of Oligosaccharyltransferase from Archaeoglobus fulgidus at 1.75 A Resolution
Biochemistry, 51, 2012
|
|
3WAK
 
 | |
3WAI
 
 | |
3WAJ
 
 | |
3VW8
 
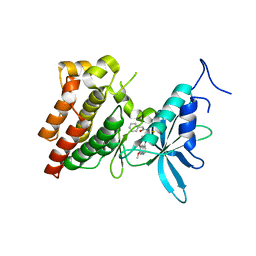 | | Crystal structure of human c-Met kinase domain with its inhibitor | | Descriptor: | CHLORIDE ION, Hepatocyte growth factor receptor, N-({4-[(6,7-dimethoxyquinolin-4-yl)oxy]phenyl}carbamothioyl)-2-phenylacetamide | | Authors: | Matsumoto, S, Miyamoto, N, Hirayama, T, Oki, H, Okada, K, Tawada, M, Iwata, H, Miki, H, Nakamura, K, Hori, A, Imamura, S. | | Deposit date: | 2012-08-08 | | Release date: | 2013-08-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design, synthesis, and evaluation of imidazo[1,2-b]pyridazine and imidazo[1,2-a]pyridine derivatives as novel dual c-Met and VEGFR2 kinase inhibitors.
Bioorg.Med.Chem., 21, 2013
|
|
4YM5
 
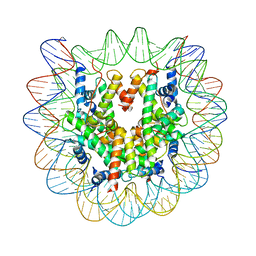 | | Crystal structure of the human nucleosome containing 6-4PP (inside) | | Descriptor: | 144 mer-DNA, 144-mer DNA, Histone H2A type 1-B/E, ... | | Authors: | Osakabe, A, Tachiwana, H, Kagawa, W, Horikoshi, N, Matsumoto, S, Hasegawa, M, Matsumoto, N, Toga, T, Yamamoto, J, Hanaoka, F, Thoma, N.H, Sugasawa, K, Iwai, S, Kurumizaka, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.005 Å) | | Cite: | Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome
Sci Rep, 5, 2015
|
|
4YM6
 
 | | Crystal structure of the human nucleosome containing 6-4PP (outside) | | Descriptor: | 145-MER DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Osakabe, A, Tachiwana, H, Kagawa, W, Horikoshi, N, Matsumoto, S, Hasegawa, M, Matsumoto, N, Toga, T, Yamamoto, J, Hanaoka, F, Thoma, N.H, Sugasawa, K, Iwai, S, Kurumizaka, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.514 Å) | | Cite: | Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome
Sci Rep, 5, 2015
|
|
8JOG
 
 | | solution structure of Ras Binding Domein (RBD) in C-RAF with negative allosteric modulator. | | Descriptor: | RAF proto-oncogene serine/threonine-protein kinase | | Authors: | Makino, Y, Matsumoto, S, Yoshikawa, Y, Kawamura, T, Kumasaka, T, Shima, F. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Small-molecule RAS/RAF-binding Inhibitors Allosterically Disrupt RAF Conformation and Exert Efficacy Against Broad-spectrum RAS-driven Cancers
To Be Published
|
|
8JOF
 
 | | solution-structure of Ras Binding Domain (RBD) in C-RAF | | Descriptor: | RAF proto-oncogene serine/threonine-protein kinase | | Authors: | Makino, Y, Matsumoto, S, Yoshikawa, Y, Kawamura, T, Kumasaka, T, Shima, F. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Small-molecule RAS/RAF-binding Inhibitors Allosterically Disrupt RAF Conformation and Exert Efficacy Against Broad-spectrum RAS-driven Cancers
To Be Published
|
|
5TNT
 
 | | Discovery of novel aminobenzisoxazole derivatives as orally available factor IXa inhibitors | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Coagulation factor IX, N-[(1S,4S,7R)-2-(3-amino-4-chloro[1,2]oxazolo[5,4-c]pyridin-7-yl)-2-azabicyclo[2.2.1]heptan-7-yl]-2-chloro-4-(3-methyl-1H-1,2,4-triazol-1-yl)benzamide, ... | | Authors: | Sakurada, I, Endo, T, Hikita, K, Hirabayashi, T, Hosaka, Y, Kato, Y, Maeda, Y, Matsumoto, S, Mizuno, T, Nagasue, A, Nishimura, T, Shimada, S, Shinozaki, M, Taguchi, K, Takeuchi, K, Yokoyama, T, Hruza, A, Reichert, P, Zhang, T, Wood, H.B, Nakao, K, Furusako, S. | | Deposit date: | 2016-10-14 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of novel aminobenzisoxazole derivatives as orally available factor IXa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5TNO
 
 | | Discovery of novel aminobenzisoxazole derivatives as orally available factor IXa inhibitors | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Coagulation factor IX, SODIUM ION, ... | | Authors: | Sakurada, I, Endo, T, Hikita, K, Hirabayashi, T, Hosaka, Y, Kato, Y, Maeda, Y, Matsumoto, S, Mizuno, T, Nagasue, H, Nishimura, T, Shimada, S, Shinozaki, M, Taguchi, K, Takeuchi, K, Yokoyama, T, Hruza, A, Reichert, P, Zhang, T, Wood, H.B, Nakao, K, Furusako, S. | | Deposit date: | 2016-10-14 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of novel aminobenzisoxazole derivatives as orally available factor IXa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7DPJ
 
 | | H-Ras Q61L in complex with GppNHp (state 1) after structural transition by humidity control | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Taniguchi, H, Matsumoto, S, Miyamoto, R, Kawamura, T, Kumasaka, T, Kataoka, T. | | Deposit date: | 2020-12-19 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.976 Å) | | Cite: | Oncogenic mutations Q61L and Q61H confer active form-like structural features to the inactive state (state 1) conformation of H-Ras protein.
Biochem.Biophys.Res.Commun., 565, 2021
|
|
7DPH
 
 | | H-Ras Q61H in complex with GppNHp (state 1) after structural transition by humidity control | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Taniguchi, H, Matsumoto, S, Kawamura, T, Kumasaka, T, Kataoka, T. | | Deposit date: | 2020-12-19 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Oncogenic mutations Q61L and Q61H confer active form-like structural features to the inactive state (state 1) conformation of H-Ras protein.
Biochem.Biophys.Res.Commun., 565, 2021
|
|
5B30
 
 | | H-Ras WT in complex with GppNHp (state 1) after structural transition by humidity control | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Kumasaka, T, Miyano, N, Baba, S, Matsumoto, S, Kataoka, T, Shima, F. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Mechanism for Conformational Dynamics of Ras-GTP Elucidated from In-Situ Structural Transition in Crystal
Sci Rep, 6, 2016
|
|
