3J15
 
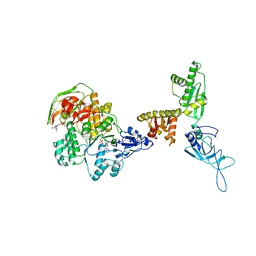 | | Model of ribosome-bound archaeal Pelota and ABCE1 | | Descriptor: | ABC transporter ATP-binding protein, ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, ... | | Authors: | Becker, T, Franckenberg, S, Wickles, S, Shoemaker, C.J, Anger, A.M, Armache, J.-P, Sieber, H, Ungewickell, C, Berninghausen, O, Daberkow, I, Karcher, A, Thomm, M, Hopfner, K.-P, Green, R, Beckmann, R. | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural basis of highly conserved ribosome recycling in eukaryotes and archaea.
Nature, 482, 2012
|
|
6I0Y
 
 | | TnaC-stalled ribosome complex with the titin I27 domain folding close to the ribosomal exit tunnel | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Su, T, Kudva, R, von Heijne, G, Beckmann, R. | | Deposit date: | 2018-10-26 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Folding pathway of an Ig domain is conserved on and off the ribosome.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3J7Z
 
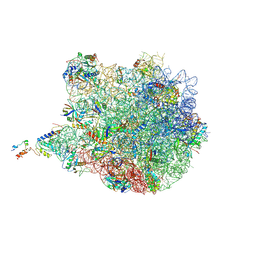 | | Structure of the E. coli 50S subunit with ErmCL nascent chain | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Arenz, S, Meydan, S, Starosta, A.L, Berninghausen, O, Beckmann, R, Vazquez-Laslop, N, Wilson, D.N. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-22 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Drug Sensing by the Ribosome Induces Translational Arrest via Active Site Perturbation.
Mol.Cell, 56, 2014
|
|
3J9Y
 
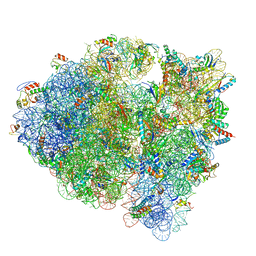 | | Cryo-EM structure of tetracycline resistance protein TetM bound to a translating E.coli ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Arenz, S, Nguyen, F, Beckmann, R, Wilson, D.N. | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-15 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the tetracycline resistance protein TetM in complex with a translating ribosome at 3.9- angstrom resolution.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3J9W
 
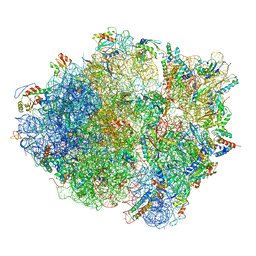 | | Cryo-EM structure of the Bacillus subtilis MifM-stalled ribosome complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein bS16, ... | | Authors: | Sohmen, D, Chiba, S, Shimokawa-Chiba, N, Innis, C.A, Berninghausen, O, Beckmann, R, Ito, K, Wilson, D.N. | | Deposit date: | 2015-03-16 | | Release date: | 2015-04-29 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the Bacillus subtilis 70S ribosome reveals the basis for species-specific stalling.
Nat Commun, 6, 2015
|
|
8AGT
 
 | | Yeast RQC complex in state F | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Tesina, P, Buschauer, R, Beckmann, R. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular basis of eIF5A-dependent CAT tailing in eukaryotic ribosome-associated quality control.
Mol.Cell, 83, 2023
|
|
8AGZ
 
 | | Yeast RQC complex in state with the RING domain of Ltn1 in the OUT position | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Tesina, P, Buschauer, R, Beckmann, R. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular basis of eIF5A-dependent CAT tailing in eukaryotic ribosome-associated quality control.
Mol.Cell, 83, 2023
|
|
8AGX
 
 | | Yeast RQC complex in state with the RING domain of Ltn1 in the IN position | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Tesina, P, Buschauer, R, Beckmann, R. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Molecular basis of eIF5A-dependent CAT tailing in eukaryotic ribosome-associated quality control.
Mol.Cell, 83, 2023
|
|
8AAF
 
 | | Yeast RQC complex in state G | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Tesina, P, Buschauer, R, Beckmann, R. | | Deposit date: | 2022-07-01 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of eIF5A-dependent CAT tailing in eukaryotic ribosome-associated quality control.
Mol.Cell, 83, 2023
|
|
8AGV
 
 | | Yeast RQC complex in state H | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Tesina, P, Buschauer, R, Beckmann, R. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular basis of eIF5A-dependent CAT tailing in eukaryotic ribosome-associated quality control.
Mol.Cell, 83, 2023
|
|
8AGW
 
 | | Yeast RQC complex in state D | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Tesina, P, Buschauer, R, Beckmann, R. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular basis of eIF5A-dependent CAT tailing in eukaryotic ribosome-associated quality control.
Mol.Cell, 83, 2023
|
|
8AGU
 
 | | Yeast RQC complex in state E | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Tesina, P, Buschauer, R, Beckmann, R. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis of eIF5A-dependent CAT tailing in eukaryotic ribosome-associated quality control.
Mol.Cell, 83, 2023
|
|
1B07
 
 | | CRK SH3 DOMAIN COMPLEXED WITH PEPTOID INHIBITOR | | Descriptor: | PHENYLETHANE, PROTEIN (PROTO-ONCOGENE CRK (CRK)), PROTEIN (SH3 PEPTOID INHIBITOR) | | Authors: | Nguyen, J.T, Turck, C.W, Cohen, F.E, Zuckermann, R.N, Lim, W.A. | | Deposit date: | 1998-11-17 | | Release date: | 1999-01-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Exploiting the basis of proline recognition by SH3 and WW domains: design of N-substituted inhibitors.
Science, 282, 1998
|
|
1JQH
 
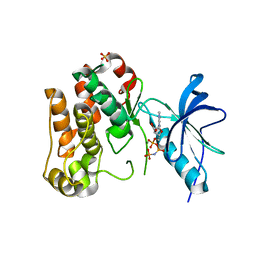 | | IGF-1 receptor kinase domain | | Descriptor: | IGF-1 receptor kinase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Pautsch, A, Zoephel, A, Ahorn, H, Spevak, W, Hauptmann, R, Nar, H. | | Deposit date: | 2001-08-07 | | Release date: | 2002-04-19 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of bisphosphorylated IGF-1 receptor kinase: insight into domain movements upon kinase activation.
Structure, 9, 2001
|
|
3J5L
 
 | | Structure of the E. coli 50S subunit with ErmBL nascent chain | | Descriptor: | 23S ribsomal RNA, 5'-R(*CP*(MA6))-3', 5'-R(*CP*CP*A)-3', ... | | Authors: | Arenz, S, Ramu, H, Gupta, P, Berninghausen, O, Beckmann, R, Vazquez-Laslop, N, Mankin, A.S, Wilson, D.N. | | Deposit date: | 2013-10-23 | | Release date: | 2014-03-26 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide.
Nat Commun, 5, 2014
|
|
8BIP
 
 | | Structure of a yeast 80S ribosome-bound N-Acetyltransferase B complex | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
8BJQ
 
 | | Structure of a yeast 80S ribosome-bound N-Acetyltransferase B complex | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-05 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
8BGU
 
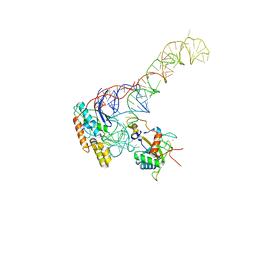 | | human MDM2-5S RNP | | Descriptor: | 5S rRNA, 60S ribosomal protein L11, 60S ribosomal protein L5, ... | | Authors: | Castillo, N, Thoms, M, Flemming, D, Hammaren, H.M, Buschauer, R, Ameismeier, M, Bassler, J, Beck, M, Beckmann, R, Hurt, E. | | Deposit date: | 2022-10-28 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of nascent 5S RNPs at the crossroad between ribosome assembly and MDM2-p53 pathways.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4PHM
 
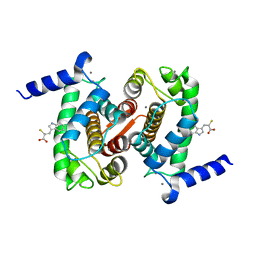 | | The Structural Basis of Differential Inhibition of Human Calpain by Indole and Phenyl alpha-Mercaptoacrylic Acids | | Descriptor: | 3-(5-bromo-1H-indol-3-yl)-2-thioxopropanoic acid, CALCIUM ION, Calpain small subunit 1 | | Authors: | Rizkallah, P.J, Allemann, R.K, Adams, S.E, Miller, D.J, Hallett, M.B. | | Deposit date: | 2014-05-06 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The structural basis of differential inhibition of human calpain by indole and phenyl alpha-mercaptoacrylic acids.
J.Struct.Biol., 187, 2014
|
|
6FTI
 
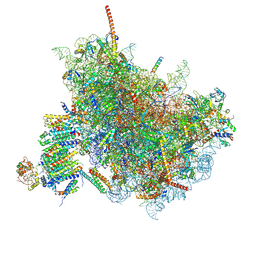 | | Cryo-EM Structure of the Mammalian Oligosaccharyltransferase Bound to Sec61 and the Programmed 80S Ribosome | | Descriptor: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Braunger, K, Becker, T, Beckmann, R. | | Deposit date: | 2018-02-22 | | Release date: | 2018-03-21 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum.
Science, 360, 2018
|
|
6FMD
 
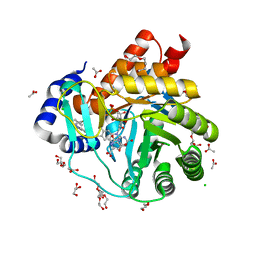 | | Targeting myeloid differentiation using potent human dihydroorotate dehydrogenase (hDHODH) inhibitors based on 2-hydroxypyrazolo[1,5-a]pyridine scaffold | | Descriptor: | 2-oxidanyl-~{N}-[2,3,5,6-tetrakis(fluoranyl)-4-phenyl-phenyl]pyrazolo[1,5-a]pyridine-3-carboxamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goyal, P, Jarva, M, Andersson, M, Lolli, M.L, Friemann, R. | | Deposit date: | 2018-01-30 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Targeting Myeloid Differentiation Using Potent 2-Hydroxypyrazolo[1,5- a]pyridine Scaffold-Based Human Dihydroorotate Dehydrogenase Inhibitors.
J. Med. Chem., 61, 2018
|
|
6G18
 
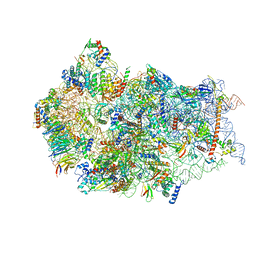 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State C | | Descriptor: | 40S ribosomal protein S11, 40S ribosomal protein S12, 40S ribosomal protein S13, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-20 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
6I7O
 
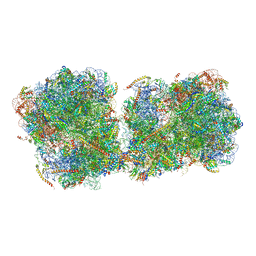 | | The structure of a di-ribosome (disome) as a unit for RQC and NGD quality control pathways recognition. | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Cheng, J, Becker, T, Beckmann, R. | | Deposit date: | 2018-11-16 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways.
EMBO J., 38, 2019
|
|
4PHJ
 
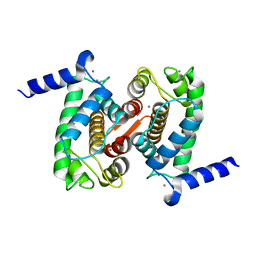 | | The Structural Basis of Differential Inhibition of Human Calpain by Indole and Phenyl alpha-Mercaptoacrylic Acids: Human unliganded protein | | Descriptor: | CALCIUM ION, Calpain small subunit 1 | | Authors: | Adams, S.E, Rizkallah, P.J, Allemann, R.K, Miller, D.J, Hallett, M.B, Robinson, E. | | Deposit date: | 2014-05-06 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural basis of differential inhibition of human calpain by indole and phenyl alpha-mercaptoacrylic acids.
J.Struct.Biol., 187, 2014
|
|
8CAH
 
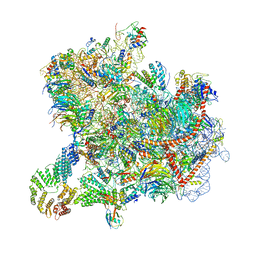 | | Cryo-EM structure of native Otu2-bound ubiquitinated 43S pre-initiation complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2023-01-24 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
