1PLG
 
 | | EVIDENCE FOR THE EXTENDED HELICAL NATURE OF POLYSACCHARIDE EPITOPES. THE 2.8 ANGSTROMS RESOLUTION STRUCTURE AND THERMODYNAMICS OF LIGAND BINDING OF AN ANTIGEN BINDING FRAGMENT SPECIFIC FOR ALPHA-(2->8)-POLYSIALIC ACID | | Descriptor: | IGG2A=KAPPA= | | Authors: | Evans, S.V, Sigurskjold, B.W, Jennings, H.J, Brisson, J.-R, Tse, W.C, To, R, Altman, E, Frosch, M, Weisgerber, C, Kratzin, H, Klebert, S, Vaesen, M, Bitter-Suermann, D, Rose, D.R, Young, N.M, Bundle, D.R. | | Deposit date: | 1995-04-24 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for the extended helical nature of polysaccharide epitopes. The 2.8 A resolution structure and thermodynamics of ligand binding of an antigen binding fragment specific for alpha-(2-->8)-polysialic acid.
Biochemistry, 34, 1995
|
|
5KSP
 
 | | hMiro1 C-domain GDP Complex C2221 Crystal Form | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Mitochondrial Rho GTPase 1 | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
2YND
 
 | | Plasmodium vivax N-myristoyltransferase in complex with a pyrazole sulphonamide inhibitor. | | Descriptor: | 2,6-dichloro-4-(2-piperazin-1-ylpyridin-4-yl)-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, 2-oxopentadecyl-CoA, CHLORIDE ION, ... | | Authors: | Wright, M.H, Clough, B, Rackham, M.D, Brannigan, J.A, Grainger, M, Bottrill, A.R, Heal, W.P, Broncel, M, Serwa, R.A, Mann, D, Leatherbarrow, R.J, Wilkinson, A.J, Holder, A.A, Tate, E.W. | | Deposit date: | 2012-10-13 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Validation of N-Myristoyltransferase as an Antimalarial Drug Target Using an Integrated Chemical Biology Approach.
Nat.Chem., 6, 2014
|
|
2YIS
 
 | | triazolopyridine inhibitors of p38 kinase. | | Descriptor: | 1-[3-tert-butyl-1-(3-chloro-4-hydroxyphenyl)-1H-pyrazol-5-yl]-3-{2-[(3-{2-[(2-hydroxyethyl)sulfanyl]phenyl}[1,2,4]triazolo[4,3-a]pyridin-6-yl)sulfanyl]benzyl}urea, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Millan, D.S, Anderson, M, Bazin, R, Bunnage, M.E, Burrows, J.L, Butcher, K.J, Dodd, P.G, Evans, T.J, Fairman, D.A, Hughes, S.J, Irving, S.L, Kilty, I.C, Lemaitre, A, Lewthwaite, R.A, Mahnke, A, Mathais, J.P, Philip, J, Phillips, C, Smith, R.T, Stefaniack, M.H, Yeadon, M. | | Deposit date: | 2011-05-16 | | Release date: | 2011-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Inhaled P38 Inhibitors for the Treatment of Chronic Obstructive Pulmonary Disease.
J.Med.Chem., 54, 2011
|
|
4RGF
 
 | | Crystal structure of the in-line aligned env22 twister ribozyme soaked with Mn2+ | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Ren, A, Rajashankar, K.R, Simanshu, D, Patel, D. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2008 Å) | | Cite: | In-line alignment and Mg(2+) coordination at the cleavage site of the env22 twister ribozyme.
Nat Commun, 5, 2014
|
|
4RGE
 
 | | Crystal structure of the in-line aligned env22 twister ribozyme | | Descriptor: | MAGNESIUM ION, env22 twister ribozyme | | Authors: | Ren, A, Rajashankar, K.R, Simanshu, D, Patel, D. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | In-line alignment and Mg(2+) coordination at the cleavage site of the env22 twister ribozyme.
Nat Commun, 5, 2014
|
|
2YNC
 
 | | Plasmodium vivax N-myristoyltransferase in complex with YnC12-CoA thioester. | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | Authors: | Wright, M.H, Clough, B, Rackham, M.D, Brannigan, J.A, Grainger, M, Bottrill, A.R, Heal, W.P, Broncel, M, Serwa, R.A, Mann, D, Leatherbarrow, R.J, Wilkinson, A.J, Holder, A.A, Tate, E.W. | | Deposit date: | 2012-10-13 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Validation of N-Myristoyltransferase as an Antimalarial Drug Target Using an Integrated Chemical Biology Approach.
Nat.Chem., 6, 2014
|
|
3N7C
 
 | | Crystal structure of the RAN binding domain from the nuclear pore complex component NUP2 from Ashbya gossypii | | Descriptor: | ABR034Wp | | Authors: | Sampathkumar, P, Manglicmot, D, Gilmore, J, Bain, K, Gheyi, T, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-26 | | Release date: | 2010-06-16 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of the RAN binding domain from the nuclear pore complex component NUP2 from Ashbya gossypii
To be Published
|
|
4RWH
 
 | | Crystal structure of T cell costimulatory ligand B7-1 (CD80) | | Descriptor: | T-lymphocyte activation antigen CD80 | | Authors: | Fedorov, A.A, Fedorov, E.V, Samanta, D, Hillerich, B, Seidel, R.D, Almo, S.C. | | Deposit date: | 2014-12-04 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal structure of T cell costimulatory ligand B7-1 (CD80)
To be Published
|
|
4RWP
 
 | | Crystal structure of porcine OAS1 in complex with dsRNA | | Descriptor: | 2'-5'-oligoadenylate synthase 1, RNA (5'-R(*GP*GP*CP*UP*UP*UP*UP*GP*AP*CP*CP*UP*UP*UP*AP*UP*GP*AP*A)-3'), RNA (5'-R(*UP*UP*CP*AP*UP*AP*AP*AP*GP*GP*UP*CP*AP*AP*AP*AP*GP*CP*C)-3') | | Authors: | Lohoefener, J, Steinke, N, Kay-Fedorov, P, Baruch, P, Nikulin, A, Tishchenko, S, Manstein, D.J, Fedorov, R. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Activation Mechanism of 2'-5'-Oligoadenylate Synthetase Gives New Insights Into OAS/cGAS Triggers of Innate Immunity.
Structure, 23, 2015
|
|
1Z64
 
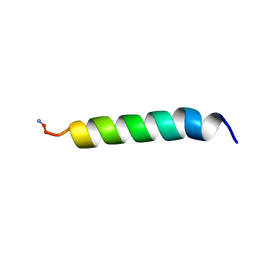 | | NMR Solution Structure of Pleurocidin in DPC Micelles | | Descriptor: | Pleruocidin | | Authors: | Syvitski, R.T, Burton, I, Mattatall, N.R, Douglas, S.E, Jakeman, D.L. | | Deposit date: | 2005-03-21 | | Release date: | 2005-04-12 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the antimicrobial peptide pleurocidin from winter flounder.
Biochemistry, 44, 2005
|
|
3N8I
 
 | |
2HVA
 
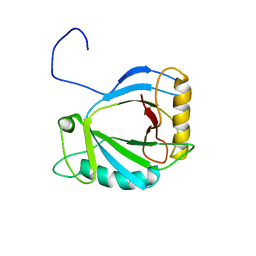 | | Solution Structure of the haem-binding protein p22HBP | | Descriptor: | Heme-binding protein 1 | | Authors: | Gell, D.A, Mackay, J.P, Westman, B.J, Liew, C.K, Gorman, D. | | Deposit date: | 2006-07-28 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A Novel Haem-binding Interface in the 22 kDa Haem-binding Protein p22HBP.
J.Mol.Biol., 362, 2006
|
|
5A3G
 
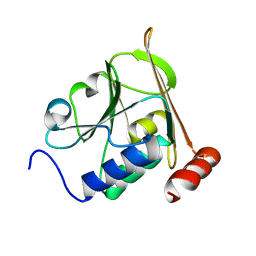 | | Structure of herpesvirus nuclear egress complex subunit M50 | | Descriptor: | M50 | | Authors: | Leigh, K.E, Boeszoermenyi, A, Mansueto, M.S, Sharma, M, Filman, D.J, Coen, D.M, Wagner, G, Hogle, J.M, Arthanari, H. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-15 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of a Herpesvirus Nuclear Egress Complex Subunit Reveals an Interaction Groove that is Essential for Viral Replication
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2Y8I
 
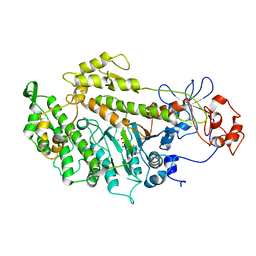 | | Structural basis for the allosteric interference of myosin function by mutants G680A and G680V of Dictyostelium myosin-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN-2 HEAVY CHAIN | | Authors: | Preller, M, Bauer, S, Adamek, N, Fujita-Becker, S, Fedorov, R, Geeves, M.A, Manstein, D.J. | | Deposit date: | 2011-02-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.132 Å) | | Cite: | Structural Basis for the Allosteric Interference of Myosin Function by Reactive Thiol Region Mutations G680A and G680V.
J.Biol.Chem., 286, 2011
|
|
4OUD
 
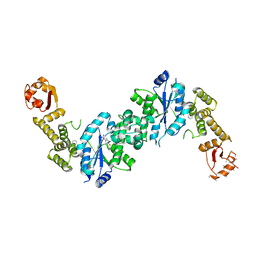 | | Engineered tyrosyl-tRNA synthetase with the nonstandard amino acid L-4,4-biphenylalanine | | Descriptor: | TYROSINE, Tyrosyl-tRNA synthetase | | Authors: | Takeuchi, R, Mandell, D.J, Lajoie, M.J, Church, G.M, Stoddard, B.L. | | Deposit date: | 2014-02-16 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biocontainment of genetically modified organisms by synthetic protein design.
Nature, 518, 2015
|
|
2WNY
 
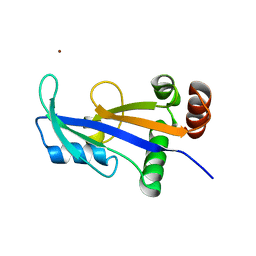 | | Structure of Mth689, a DUF54 protein from Methanothermobacter thermautotrophicus | | Descriptor: | CHLORIDE ION, CONSERVED PROTEIN MTH689, NICKEL (II) ION | | Authors: | Ng, C.L, Waterman, D.G, Lebedev, A.A, Smits, C, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2009-07-21 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Mth689, a Duf54 Protein from Methanothermobacter Thermautotrophicus
To be Published
|
|
2X9H
 
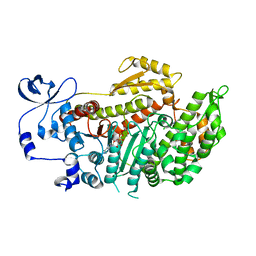 | | CRYSTAL STRUCTURE OF MYOSIN-2 MOTOR DOMAIN IN COMPLEX WITH ADP- METAVANADATE AND PENTACHLOROCARBAZOLE | | Descriptor: | 2,3,4,6,8-PENTACHLORO-9H-CARBAZOL-1-OL, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Selvadurai, J, Kirst, J, Knoelker, H.J, Manstein, D.J. | | Deposit date: | 2010-03-19 | | Release date: | 2011-05-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Myosin-2 Motor Domain in Complex with Adp-Metavanadate and Pentachlorocarbazole
To be Published
|
|
4LRB
 
 | | Phosphopentomutase S154G variant soaked with 2,3-dideoxyribose 5-phosphate | | Descriptor: | 2,3-dideoxy-5-O-phosphono-alpha-D-ribofuranose, ACETATE ION, GLYCEROL, ... | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
4LR8
 
 | | Phosphopentomutase S154A variant soaked with ribose 5-phosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-O-phosphono-alpha-D-ribofuranose, ACETATE ION, ... | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
3J9F
 
 | | Poliovirus complexed with soluble, deglycosylated poliovirus receptor (Pvr) at 4 degrees C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Strauss, M, Filman, D.J, Belnap, D.M, Cheng, N, Noel, R.T, Hogle, J.M. | | Deposit date: | 2015-01-15 | | Release date: | 2015-02-11 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Nectin-Like Interactions between Poliovirus and Its Receptor Trigger Conformational Changes Associated with Cell Entry.
J.Virol., 89, 2015
|
|
4LRE
 
 | | Phosphopentomutase soaked with 2,3-dideoxyribose 5-phosphate | | Descriptor: | 2,3-dideoxy-5-O-phosphono-alpha-D-ribofuranose, MANGANESE (II) ION, Phosphopentomutase | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
2JJ9
 
 | | Crystal structure of myosin-2 in complex with ADP-metavanadate | | Descriptor: | ADP METAVANADATE, MAGNESIUM ION, MYOSIN-2 HEAVY CHAIN | | Authors: | Fedorov, R, Boehl, M, Tsiavaliaris, G, Hartmann, F.K, Baruch, P, Brenner, B, Martin, R, Knoelker, H.J, Gutzeit, H.O, Manstein, D.J. | | Deposit date: | 2008-03-25 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Mechanism of Pentabromopseudilin Inhibition of Myosin Motor Activity.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2ZVO
 
 | | NEMO CoZi domain in complex with diubiquitin in C2 space group | | Descriptor: | NF-kappa-B essential modulator, UBC protein | | Authors: | Rahighi, S, Ikeda, F, Kawasaki, M, Akutsu, M, Suzuki, N, Kato, R, Kensche, T, Uejima, T, Bloor, S, Komander, D, Randow, F, Wakatsuki, S, Dikic, I. | | Deposit date: | 2008-11-12 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation
Cell(Cambridge,Mass.), 136, 2009
|
|
4LR9
 
 | | Phosphopentomutase S154A variant soaked with 2,3-dideoxyribose 5-phosphate | | Descriptor: | 2,3-dideoxy-5-O-phosphono-alpha-D-ribofuranose, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
