8DM7
 
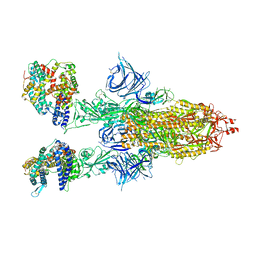 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DM5
 
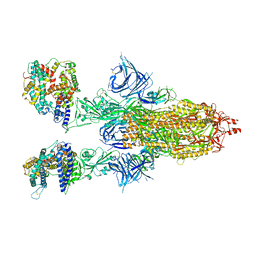 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
4R0E
 
 | |
5JK4
 
 | | Phosphate-Binding Protein from Stenotrophomonas maltophilia. | | Descriptor: | Alkaline phosphatase, PHOSPHATE ION | | Authors: | Keegan, R, Waterman, D, Hopper, D, Coates, L, Guo, J, Coker, A.R, Erskine, P.T, Wood, S.P, Cooper, J.B. | | Deposit date: | 2016-04-25 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1 angstrom resolution structure of a periplasmic phosphate-binding protein from Stenotrophomonas maltophilia: a crystallization contaminant identified by molecular replacement using the entire Protein Data Bank.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3M8W
 
 | | Phosphopentomutase from Bacillus cereus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Panosian, T.D, Nannemann, D.P, Watkins, G, Wadzinski, B, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2010-03-19 | | Release date: | 2010-12-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bacillus cereus Phosphopentomutase Is an Alkaline Phosphatase Family Member That Exhibits an Altered Entry Point into the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
3M8Y
 
 | | Phosphopentomutase from Bacillus cereus after glucose-1,6-bisphosphate activation | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Panosian, T.D, Nannemann, D.P, Watkins, G, Wadzinski, B, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2010-03-19 | | Release date: | 2010-12-29 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacillus cereus Phosphopentomutase Is an Alkaline Phosphatase Family Member That Exhibits an Altered Entry Point into the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
5KWW
 
 | | Crystal Structure of Inhibitor JNJ-53718678 In Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[[5-chloranyl-1-(3-methylsulfonylpropyl)indol-2-yl]methyl]-1-[2,2,2-tris(fluoranyl)ethyl]imidazo[4,5-c]pyridin-2-one, ... | | Authors: | McLellan, J.S, Battles, M.B, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2016-07-19 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Therapeutic efficacy of a respiratory syncytial virus fusion inhibitor.
Nat Commun, 8, 2017
|
|
5KNQ
 
 | | Crystal structure of Mycobacterium tuberculosis hypoxanthine guanine phosphoribosyltransferase in complex with [3S,4R]-(4-(Guanin-9-yl)pyrrolidin-3-yl)oxymethanephosphonic acid and pyrophosphate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Eng, W.S, Rejman, D, Keough, D.T, Guddat, L.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-09-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis hypoxanthine guanine phosphoribosyltransferase in complex with pyrrolidine nucleoside phosphonate
To Be Published
|
|
5JP3
 
 | |
3NF5
 
 | | Crystal structure of the C-terminal domain of nuclear pore complex component NUP116 from Candida glabrata | | Descriptor: | GLYCEROL, Nucleoporin NUP116 | | Authors: | Sampathkumar, P, Manglicmot, D, Bain, K, Gilmore, J, Gheyi, T, Rout, M, Sali, A, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata.
Proteins, 80, 2012
|
|
5KNY
 
 | | Crystal structure of Mycobacterium tuberculosis hypoxanthine guanine phosphoribosyltransferase in complex with (3-((3R,4R)-3-(Guanin-9-yl)-4-((S)-2-hydroxy-2-phosphonoethoxy)pyrrolidin-1-yl)-3-oxopropyl)phosphonic acid | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION, [3-[(3~{R},4~{R})-3-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-4-[(2~{S})-2-oxidanyl-2-phosphono-ethoxy]pyrrolidin-1-y l]-3-oxidanylidene-propyl]phosphonic acid | | Authors: | Eng, W.S, Rejman, D, Keough, D.T, Guddat, L.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis hypoxanthine guanine phosphoribosyltransferase in complex with pyrrolidine nucleoside phosphonate
To Be Published
|
|
5WPS
 
 | | Crystal structure HpiC1 Y101F | | Descriptor: | 1,2-ETHANEDIOL, 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.389 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
4S14
 
 | | Crystal structure of the orphan nuclear receptor RORgamma ligand-binding domain in complex with 4alpha-caboxyl, 4beta-methyl-zymosterol (4ACD8) | | Descriptor: | (3beta,4alpha,5beta,14beta)-3-hydroxy-4-methylcholesta-8,24-diene-4-carboxylic acid, Nuclear receptor ROR-gamma, Nuclear receptor-interacting protein 1 | | Authors: | Huang, P, Santori, F.R, Littman, D.R, Rastinejad, F. | | Deposit date: | 2015-01-07 | | Release date: | 2015-02-11 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (3.542 Å) | | Cite: | Identification of Natural ROR gamma Ligands that Regulate the Development of Lymphoid Cells.
Cell Metab, 21, 2015
|
|
5IFR
 
 | | Structure of the stable UBE2D3-UbDha conjugate | | Descriptor: | GLYCEROL, Polyubiquitin-B, Ubiquitin-conjugating enzyme E2 D3 | | Authors: | Pruneda, J.N, Mulder, M.P.C, Witting, K, Ovaa, H, Komander, D. | | Deposit date: | 2016-02-26 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A cascading activity-based probe sequentially targets E1-E2-E3 ubiquitin enzymes.
Nat.Chem.Biol., 12, 2016
|
|
8E34
 
 | | CryoEM structures of bAE1 captured in multiple states | | Descriptor: | Anion exchange protein | | Authors: | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2022-08-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
2YGX
 
 | | Structure of the mixed-function P450 MycG in P21 space group | | Descriptor: | GLYCEROL, P-450-LIKE PROTEIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Li, S, Kells, P.M, Rutaganira, F.U, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-04-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
4OYJ
 
 | | Structure of the apo HOIP PUB domain | | Descriptor: | E3 ubiquitin-protein ligase RNF31, SULFATE ION | | Authors: | Elliott, P.R, Komander, D. | | Deposit date: | 2014-02-12 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular Basis and Regulation of OTULIN-LUBAC Interaction.
Mol.Cell, 54, 2014
|
|
2YIW
 
 | | triazolopyridine inhibitors of p38 kinase | | Descriptor: | 1-(3-tert-butyl-1-phenyl-1H-pyrazol-5-yl)-3-(2-{[3-(1-methylethyl)[1,2,4]triazolo[4,3-a]pyridin-6-yl]sulfanyl}benzyl)urea, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Millan, D.S, Anderson, M, Bunnage, M.E, Burrows, J.L, Butcher, K.J, Dodd, P.G, Evans, T.J, Fairman, D.A, Hughes, S.J, Irving, S.L, Kilty, I.C, Lemaitre, A, Lewthwaite, R.A, Mahnke, A, Mathais, J.P, Philip, J, Phillips, C, Smith, R.T, Stefamiak, M.H, Yeadon, M. | | Deposit date: | 2011-05-17 | | Release date: | 2011-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Inhaled P38 Inhibitors for the Treatment of Chronic Obstructive Pulmonary Disease.
J.Med.Chem., 54, 2011
|
|
5B3I
 
 | | Homo-dimeric structure of cytochrome c' from Thermophilic Hydrogenophilus thermoluteolus | | Descriptor: | Cytochrome c prime, HEME C | | Authors: | Fujii, S, Oki, H, Kawahara, K, Yamane, D, Yamanaka, M, Maruno, T, Kobayashi, Y, Masanari, M, Wakai, S, Nishihara, H, Ohkubo, T, Sambongi, Y. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and functional insights into thermally stable cytochrome c' from a thermophile
Protein Sci., 26, 2017
|
|
2YNE
 
 | | Plasmodium vivax N-myristoyltransferase in complex with a benzothiophene inhibitor | | Descriptor: | 2-(3-piperidin-4-yloxy-1-benzothiophen-2-yl)-5-[(1,3,5-trimethylpyrazol-4-yl)methyl]-1,3,4-oxadiazole, 2-oxopentadecyl-CoA, CHLORIDE ION, ... | | Authors: | Wright, M.H, Clough, B, Rackham, M.D, Brannigan, J.A, Grainger, M, Bottrill, A.R, Heal, W.P, Broncel, M, Serwa, R.A, Mann, D, Leatherbarrow, R.J, Wilkinson, A.J, Holder, A.A, Tate, E.W. | | Deposit date: | 2012-10-13 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Validation of N-Myristoyltransferase as an Antimalarial Drug Target Using an Integrated Chemical Biology Approach.
Nat.Chem., 6, 2014
|
|
2Z6R
 
 | | Crystal structure of Lys49 to Arg mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Krishna Swamy, B.S, Simanshu, D.K, Murthy, M.R.N, Kunishima, N. | | Deposit date: | 2007-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
2WNR
 
 | | The structure of Methanothermobacter thermautotrophicus exosome core assembly | | Descriptor: | PHOSPHATE ION, PROBABLE EXOSOME COMPLEX EXONUCLEASE 1, PROBABLE EXOSOME COMPLEX EXONUCLEASE 2 | | Authors: | Ng, C.L, Waterman, D.G, Antson, A.A, Ortiz-Lombardia, M. | | Deposit date: | 2009-07-19 | | Release date: | 2010-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the Methanothermobacter Thermautotrophicus Exosome Rnase Ph Ring
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4OYK
 
 | |
5JLH
 
 | | Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 2, ... | | Authors: | von der Ecken, J, Heissler, S.M, Pathan-Chhatbar, S, Manstein, D.J, Raunser, S. | | Deposit date: | 2016-04-27 | | Release date: | 2016-06-15 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution.
Nature, 534, 2016
|
|
5J2U
 
 | | Tubulin-MMAF complex | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Waight, A.B, Bargsten, K, Doronina, S, Steinmetz, M.O, Sussman, D, Prota, A.E. | | Deposit date: | 2016-03-30 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Microtubule Destabilization by Potent Auristatin Anti-Mitotics.
Plos One, 11, 2016
|
|
