7UO4
 
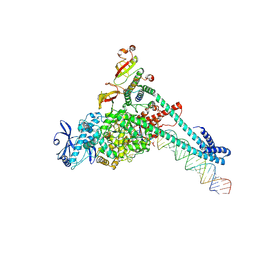 | | SARS-CoV-2 replication-transcription complex bound to Remdesivir triphosphate, in a pre-catalytic state | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UOB
 
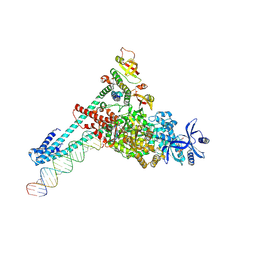 | | SARS-CoV-2 replication-transcription complex bound to GTP, in a pre-catalytic state | | Descriptor: | 3'-DEOXYURIDINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UO9
 
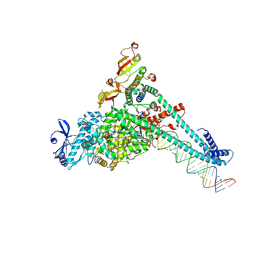 | | SARS-CoV-2 replication-transcription complex bound to UTP, in a pre-catalytic state | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UO7
 
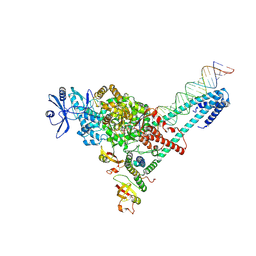 | | SARS-CoV-2 replication-transcription complex bound to ATP, in a pre-catalytic state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UOE
 
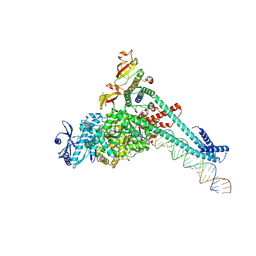 | | SARS-CoV-2 replication-transcription complex bound to CTP, in a pre-catalytic state | | Descriptor: | 3'-DEOXYURIDINE-5'-MONOPHOSPHATE, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7RDX
 
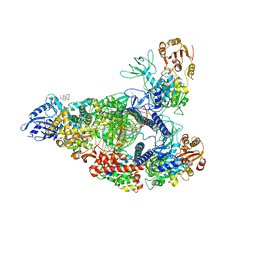 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - open class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RDZ
 
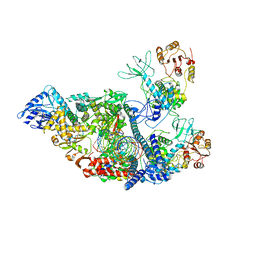 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - apo class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE3
 
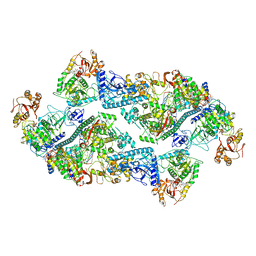 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RDY
 
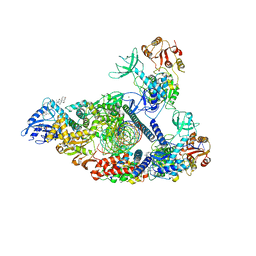 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - engaged class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE0
 
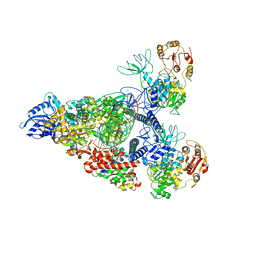 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - swiveled class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE2
 
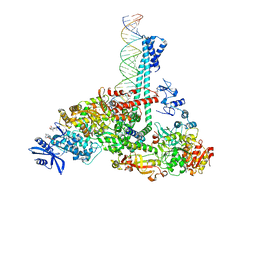 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(1)-RTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE1
 
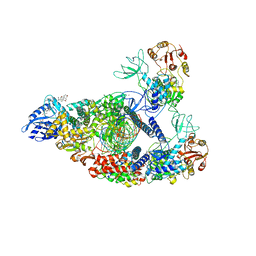 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC (composite) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7KRN
 
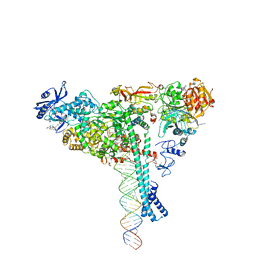 | | Structure of SARS-CoV-2 backtracked complex bound to nsp13 helicase - nsp13(1)-BTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KRP
 
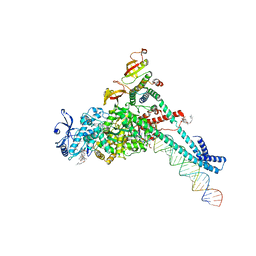 | | Structure of SARS-CoV-2 backtracked complex complex bound to nsp13 helicase - BTC (local refinement) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHAPSO, MAGNESIUM ION, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KRO
 
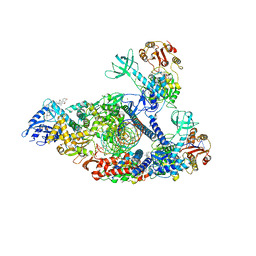 | | Structure of SARS-CoV-2 backtracked complex complex bound to nsp13 helicase - nsp13(2)-BTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6XEZ
 
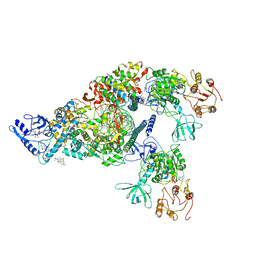 | | Structure of SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Llewellyn, E.C, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex.
Cell, 182, 2020
|
|
6Y8N
 
 | | Mycobacterium thermoresistibile GyrB21 in complex with Redx03863 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(1~{S},5~{R})-6-azanyl-3-azabicyclo[3.1.0]hexan-3-yl]-6-fluoranyl-~{N}-methyl-2-(2-methylpyrimidin-5-yl)oxy-9~{H}-pyrimido[4,5-b]indol-8-amine, DNA gyrase subunit B, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
6Y8L
 
 | | Mycobacterium thermoresistibile GyrB21 in complex with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, NOVOBIOCIN, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
6Y8O
 
 | | Mycobacterium smegmatis GyrB 22kDa ATPase sub-domain in complex with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA gyrase subunit B, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
