1M3V
 
 | | FLIN4: Fusion of the LIM binding domain of Ldb1 and the N-terminal LIM domain of LMO4 | | Descriptor: | ZINC ION, fusion of the LIM interacting domain of ldb1 and the N-terminal LIM domain of LMO4 | | Authors: | Deane, J.E, Mackay, J.P, Kwan, A.H.Y, Sum, E.Y, Visvader, J.E, Matthews, J.M. | | Deposit date: | 2002-06-30 | | Release date: | 2003-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of ldb1 by the N-terminal LIM domains of LMO2 and LMO4
EMBO J., 22, 2003
|
|
6BGG
 
 | | Solution NMR structures of the BRD3 ET domain in complex with a CHD4 peptide | | Descriptor: | Bromodomain-containing protein 3, CHD4 | | Authors: | Wai, D.C.C, Szyszka, T.N, Campbell, A.E, Kwong, C, Wilkinson-White, L, Silva, A.P.G, Low, J.K.K, Kwan, A.H, Gamsjaeger, R, Lu, B, Vakoc, C.R, Blobel, G.A, Mackay, J.P. | | Deposit date: | 2017-10-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The BRD3 ET domain recognizes a short peptide motif through a mechanism that is conserved across chromatin remodelers and transcriptional regulators.
J. Biol. Chem., 293, 2018
|
|
6BI6
 
 | | Solution NMR structure of uncharacterized protein YejG | | Descriptor: | Uncharacterized protein YejG | | Authors: | Mohanty, B, Finn, T.J, Macindoe, I, Zhong, J, Patrick, W.M, Mackay, J.P. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The uncharacterized bacterial protein YejG has the same architecture as domain III of elongation factor G.
Proteins, 87, 2019
|
|
6BGH
 
 | |
8D4Y
 
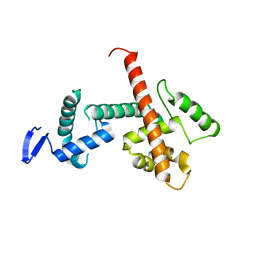 | | C-terminal SANT-SLIDE domain of human Chromodomain-helicase-DNA-binding protein 4 (CHD4) | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4 | | Authors: | Moghaddas Sani, H, Deshpande, C.N, Panjikar, S, Patel, K, Mackay, J.P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The role of auxiliary domains in modulating CHD4 activity suggests mechanistic commonality between enzyme families.
Nat Commun, 13, 2022
|
|
8CV5
 
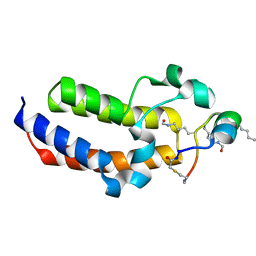 | | Peptide 4.2B in complex with BRD3.2 | | Descriptor: | ACETYL GROUP, AMINO GROUP, Bromodomain-containing protein 3, ... | | Authors: | Franck, C, Mackay, J.P. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery and characterization of cyclic peptides selective for the C-terminal bromodomains of BET family proteins.
Structure, 31, 2023
|
|
8CV7
 
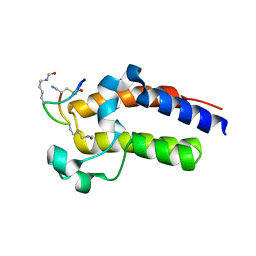 | | Peptide 2.2E in complex with BRD2-BD2 | | Descriptor: | ACETYL GROUP, AMINO GROUP, Isoform 3 of Bromodomain-containing protein 2, ... | | Authors: | Franck, C, Mackay, J.P. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and characterization of cyclic peptides selective for the C-terminal bromodomains of BET family proteins.
Structure, 31, 2023
|
|
8CV6
 
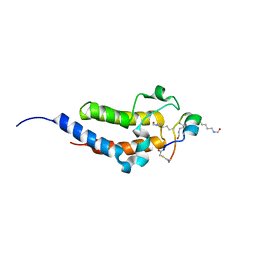 | | Peptide 4.2B in complex with BRD4.2 | | Descriptor: | ACETYL GROUP, AMINO GROUP, BRD4 protein, ... | | Authors: | Franck, C, Mackay, J.P. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and characterization of cyclic peptides selective for the C-terminal bromodomains of BET family proteins.
Structure, 31, 2023
|
|
8CV4
 
 | | Peptide 4.2C in complex with BRD4.2 | | Descriptor: | ACETYL GROUP, AMINO GROUP, BRD4 protein, ... | | Authors: | Franck, C, Mackay, J.P. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery and characterization of cyclic peptides selective for the C-terminal bromodomains of BET family proteins.
Structure, 31, 2023
|
|
4PBZ
 
 | | Structure of the human RbAp48-MTA1(670-695) complex | | Descriptor: | Histone-binding protein RBBP4, Metastasis-associated protein MTA1 | | Authors: | Murthy, A, Pei, X.Y, Watson, A.A, Silva, A.P.G, Mackay, J.P, Laue, E.D. | | Deposit date: | 2014-04-14 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insight into the architecture of the NuRD complex: Structure of the RbAp48-MTA1 sub-complex.
J.Biol.Chem., 289, 2014
|
|
4PBY
 
 | | Structure of the human RbAp48-MTA1(656-686) complex | | Descriptor: | Histone-binding protein RBBP4, ISOPROPYL ALCOHOL, Metastasis-associated protein MTA1 | | Authors: | Murthy, A, Lejon, S, Alqarni, S.S.M, Silva, A.P.G, Watson, A.A, Mackay, J.P, Laue, E.D. | | Deposit date: | 2014-04-14 | | Release date: | 2014-06-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into the architecture of the NuRD complex: Structure of the RbAp48-MTA1 sub-complex.
J.Biol.Chem., 289, 2014
|
|
4PC0
 
 | | Structure of the human RbAp48-MTA1(670-711) complex | | Descriptor: | CALCIUM ION, GLYCEROL, Histone-binding protein RBBP4, ... | | Authors: | Alqarni, S.S.M, Silva, A.P.G, Mackay, J.P, Laue, E.D. | | Deposit date: | 2014-04-14 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into the architecture of the NuRD complex: Structure of the RbAp48-MTA1 sub-complex.
J.Biol.Chem., 289, 2014
|
|
8DNQ
 
 | | BRD2-BD1 in complex with cyclic peptide 2.2B | | Descriptor: | Bromodomain-containing protein 2, Cyclic peptide 2.2B, GLYCEROL | | Authors: | Patel, K, Franck, C, Mackay, J.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-07-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery and characterization of cyclic peptides selective for the C-terminal bromodomains of BET family proteins.
Structure, 31, 2023
|
|
4F2J
 
 | | Crystal structure of ZNF217 bound to DNA, P6522 crystal form | | Descriptor: | 5'-D(*TP*TP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', ZINC ION, Zinc finger protein 217 | | Authors: | Vandevenne, M.S, Jacques, D.A, Guss, J.M, Mackay, J.P. | | Deposit date: | 2012-05-08 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | New insights into DNA recognition by zinc fingers revealed by structural analysis of the oncoprotein ZNF217
J.Biol.Chem., 288, 2013
|
|
1J2O
 
 | | Structure of FLIN2, a complex containing the N-terminal LIM domain of LMO2 and ldb1-LID | | Descriptor: | Fusion of Rhombotin-2 and LIM domain-binding protein 1, ZINC ION | | Authors: | Deane, J.E, Mackay, J.P, Kwan, A.H, Sum, E.Y, Visvader, J.E, Matthews, J.M. | | Deposit date: | 2003-01-08 | | Release date: | 2003-05-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of ldb1 by the N-terminal LIM domains of LMO2 and LMO4
EMBO J., 22, 2003
|
|
1FV5
 
 | | SOLUTION STRUCTURE OF THE FIRST ZINC FINGER FROM THE DROSOPHILA U-SHAPED TRANSCRIPTION FACTOR | | Descriptor: | FIRST ZINC FINGER OF U-SHAPED, ZINC ION | | Authors: | Liew, C.K, Kowalski, K, Fox, A.H, Newton, A, Sharpe, B.K, Crossley, M, Mackay, J.P. | | Deposit date: | 2000-09-18 | | Release date: | 2000-10-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two CCHC zinc fingers from the FOG family protein U-shaped that mediate protein-protein interactions.
Structure Fold.Des., 8, 2000
|
|
1FU9
 
 | | SOLUTION STRUCTURE OF THE NINTH ZINC-FINGER DOMAIN OF THE U-SHAPED TRANSCRIPTION FACTOR | | Descriptor: | U-SHAPED TRANSCRIPTIONAL COFACTOR, ZINC ION | | Authors: | Liew, C.K, Kowalski, K, Fox, A.H, Newton, A, Sharpe, B.K, Crossley, M, Mackay, J.P. | | Deposit date: | 2000-09-14 | | Release date: | 2000-10-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two CCHC zinc fingers from the FOG family protein U-shaped that mediate protein-protein interactions.
Structure Fold.Des., 8, 2000
|
|
1GNF
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZINC FINGER OF MURINE GATA-1, NMR, 25 STRUCTURES | | Descriptor: | TRANSCRIPTION FACTOR GATA-1, ZINC ION | | Authors: | Kowalski, K, Czolij, R, King, G.F, Crossley, M, Mackay, J.P. | | Deposit date: | 1998-10-12 | | Release date: | 1999-06-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal zinc finger of GATA-1 reveals a specific binding face for the transcriptional co-factor FOG.
J.Biomol.NMR, 13, 1999
|
|
1P7A
 
 | | Solution Structure of the Third Zinc Finger from BKLF | | Descriptor: | Kruppel-like factor 3, ZINC ION | | Authors: | Simpson, R.J.Y, Cram, E.D, Czolij, R, Matthews, J.M, Crossley, M, Mackay, J.P. | | Deposit date: | 2003-04-30 | | Release date: | 2003-12-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | CCHX zinc finger derivatives retain the ability to bind Zn(II) and mediate protein-DNA interactions.
J.Biol.Chem., 278, 2003
|
|
1N0Z
 
 | | Solution structure of the first zinc-finger domain from ZNF265 | | Descriptor: | ZINC ION, ZNF265 | | Authors: | Plambeck, C.A, Fairley, K, Kwan, A.H.Y, Westman, B.J, Adams, D, Morris, B, Mackay, J.P. | | Deposit date: | 2002-10-15 | | Release date: | 2003-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of the zinc finger domain from human splicing factor ZNF265 fold
J.BIOL.CHEM., 278, 2003
|
|
1MM2
 
 | | Solution structure of the 2nd PHD domain from Mi2b | | Descriptor: | Mi2-beta, ZINC ION | | Authors: | Kwan, A.H.Y, Gell, D.A, Verger, A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2002-09-02 | | Release date: | 2003-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Engineering a Protein Scaffold from a PHD Finger
structure, 11, 2003
|
|
1PM4
 
 | | Crystal structure of Yersinia pseudotuberculosis-derived mitogen (YPM) | | Descriptor: | YPM | | Authors: | Donadini, R, Liew, C.W, Kwan, A.H, Mackay, J.P, Fields, B.A. | | Deposit date: | 2003-06-09 | | Release date: | 2004-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Crystal and Solution Structures of a Superantigen from Yersinia pseudotuberculosis Reveal a Jelly-Roll Fold.
Structure, 12, 2004
|
|
1POQ
 
 | | Solution Structure of a Superantigen from Yersinia pseudotuberculosis | | Descriptor: | YPM | | Authors: | Donadini, R, Liew, C.W, Kwan, A.H, Mackay, J.P, Fields, B.A. | | Deposit date: | 2003-06-16 | | Release date: | 2004-01-27 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Crystal and Solution Structures of a Superantigen from Yersinia pseudotuberculosis Reveal a Jelly-Roll Fold.
Structure, 12, 2004
|
|
1K2F
 
 | | siah, Seven In Absentia Homolog | | Descriptor: | BETA-MERCAPTOETHANOL, ZINC ION, siah-1A protein | | Authors: | Polekhina, G, House, C.M, Traficante, N, Mackay, J.P, Relaix, F, Sassoon, D.A, Parker, M.W, Bowtell, D.D.L. | | Deposit date: | 2001-09-26 | | Release date: | 2001-12-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-alpha signaling.
Nat.Struct.Biol., 9, 2002
|
|
1JN7
 
 | |
