6JWA
 
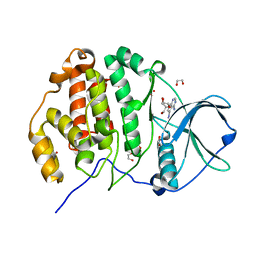 | | Crystal structure of CK2a1 with 5-iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, Casein kinase II subunit alpha | | Authors: | Tsuyuguchi, M, Kinoshita, T. | | Deposit date: | 2019-04-19 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | A promiscuous kinase inhibitor delineates the conspicuous structural features of protein kinase CK2a1.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
2PCF
 
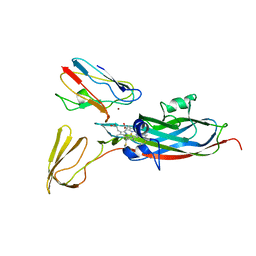 | | THE COMPLEX OF CYTOCHROME F AND PLASTOCYANIN DETERMINED WITH PARAMAGNETIC NMR. BASED ON THE STRUCTURES OF CYTOCHROME F AND PLASTOCYANIN, 10 STRUCTURES | | Descriptor: | COPPER (II) ION, CYTOCHROME F, HEME C, ... | | Authors: | Ubbink, M, Ejdeback, M, Karlsson, B.G, Bendall, D.S. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The structure of the complex of plastocyanin and cytochrome f, determined by paramagnetic NMR and restrained rigid-body molecular dynamics.
Structure, 6, 1998
|
|
1K37
 
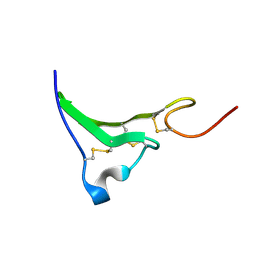 | | NMR Structure of human Epiregulin | | Descriptor: | Epiregulin | | Authors: | Sato, K, Miura, K, Tada, M, Aizawa, T, Miyamoto, K, Kawano, K. | | Deposit date: | 2001-10-02 | | Release date: | 2003-09-30 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of epiregulin and the effect of its C-terminal domain for receptor binding affinity
Febs Lett., 553, 2003
|
|
3RZP
 
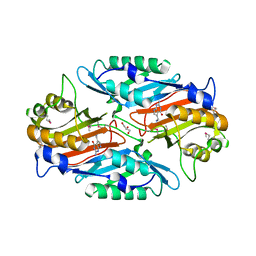 | | Crystal Structure of the C194A mutant of 7-cyano-7-deazaguanine reductase, QueF from Vibrio cholerae complexed with preQ1 | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, GLYCEROL, NADPH-dependent 7-cyano-7-deazaguanine reductase | | Authors: | Kim, Y, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-12 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the C194A mutant of 7-cyano-7-deazaguanine reductase, QueF from Vibrio cholerae complexed with preQ1
To be Published
|
|
7DBS
 
 | |
1K36
 
 | | NMR Structure of human Epiregulin | | Descriptor: | Epiregulin | | Authors: | Sato, K, Miura, K, Tada, M, Aizawa, T, Miyamoto, K, Kawano, K. | | Deposit date: | 2001-10-02 | | Release date: | 2003-09-30 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of epiregulin and the effect of its C-terminal domain for receptor binding affinity
Febs Lett., 553, 2003
|
|
3B21
 
 | |
4LL3
 
 | | Structure of wild-type HIV protease in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Grantz Saskova, K, Rezacova, P, Brynda, J, Kozisek, M, Konvalinka, J. | | Deposit date: | 2013-07-09 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Thermodynamic and structural analysis of HIV protease resistance to darunavir - analysis of heavily mutated patient-derived HIV-1 proteases.
Febs J., 281, 2014
|
|
1TIV
 
 | |
1TBC
 
 | |
3AB3
 
 | | Crystal structure of p115RhoGEF RGS domain in complex with G alpha 13 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(k) subunit alpha, Guanine nucleotide-binding protein subunit alpha-13, ... | | Authors: | Kukimoto-Niino, M, Mishima, C, Shirouzu, M, Kozasa, T, Yokoyama, S. | | Deposit date: | 2009-11-30 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of critical residues in G(alpha)13 for stimulation of p115RhoGEF activity and the structure of the G(alpha)13-p115RhoGEF regulator of G protein signaling homology (RH) domain complex.
J.Biol.Chem., 286, 2011
|
|
6KJK
 
 | | Structure of the N-terminal domain of PorA | | Descriptor: | N-terminal domain of PorA (28-171) | | Authors: | Handa, Y, Sato, K, Shoji, M, Nakayama, K, Imada, K. | | Deposit date: | 2019-07-22 | | Release date: | 2020-07-22 | | Last modified: | 2020-12-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PorA, a conserved C-terminal domain-containing protein, impacts the PorXY-SigP signaling of the type IX secretion system.
Sci Rep, 10, 2020
|
|
3VJT
 
 | | Vitamin D receptor complex with a carborane compound | | Descriptor: | 1-(2-[(R)-2,4-Dihydroxybutoxy]ethyl)-12-(5-ethyl-5-hydroxyheptyl)-1,12-dicarba-closo-dodecaborane, Vitamin D3 receptor, peptide from Mediator of RNA polymerase II transcription subunit 1 | | Authors: | Fujii, S, Masuno, M, Kagechika, H, Nakabayashi, M, Ito, N. | | Deposit date: | 2011-10-31 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Boron Cluster-based Development of Potent Nonsecosteroidal Vitamin D Receptor Ligands: Direct Observation of Hydrophobic Interaction between Protein Surface and Carborane
J.Am.Chem.Soc., 133, 2011
|
|
3AE7
 
 | | Crystal structure of porcine heart mitochondrial complex II bound with 2-Iodo-N-(3-isopropoxy-phenyl)-benzamide | | Descriptor: | 2-iodo-N-[3-(1-methylethoxy)phenyl]benzamide, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Sasaki, T, Shindo, M, Kido, Y, Inaoka, D.K, Omori, J, Osanai, A, Sakamoto, K, Mao, J, Matsuoka, S, Inoue, M, Honma, T, Tanaka, A, Kita, K. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
3AE9
 
 | | Crystal structure of porcine heart mitochondrial complex II bound with N-(3-Pentafluorophenyloxy-phenyl)-2-trifluoromethyl-benzamide | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Harada, S, Sasaki, T, Shindo, M, Kido, Y, Inaoka, D.K, Omori, J, Osanai, A, Sakamoto, K, Mao, J, Matsuoka, S, Inoue, M, Honma, T, Tanaka, A, Kita, K. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
3VMP
 
 | | Crystal structure of dextranase from Streptococcus mutans in complex with 4,5-epoxypentyl alpha-D-glucopyranoside | | Descriptor: | 5-hydroxypentyl alpha-D-glucopyranoside, Dextranase, PHOSPHATE ION | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Okuyama, M, Mori, H, Funane, K, Kimura, A. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural elucidation of dextran degradation mechanism by streptococcus mutans dextranase belonging to glycoside hydrolase family 66
J.Biol.Chem., 287, 2012
|
|
4PTE
 
 | | Structure of a carvoxamide compound (15) (N-[4-(ISOQUINOLIN-7-YL)PYRIDIN-2-YL]CYCLOPROPANECARBOXAMIDE) to GSK3b | | Descriptor: | Glycogen synthase kinase-3 beta, N-[4-(isoquinolin-7-yl)pyridin-2-yl]cyclopropanecarboxamide | | Authors: | Lewis, H.A, Sivaprakasam, P, Kish, K, Pokross, M, Dubowchik, G.M. | | Deposit date: | 2014-03-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Discovery of new acylaminopyridines as GSK-3 inhibitors by a structure guided in-depth exploration of chemical space around a pyrrolopyridinone core.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6JZQ
 
 | |
3APC
 
 | | Crystal structure of human PI3K-gamma in complex with CH5132799 | | Descriptor: | 5-(7-Methanesulfonyl-2-morpholin-4-yl-6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidin-4-yl)-pyrimidin-2-ylamine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Nakamura, M, Fukami, T.A, Miyazaki, T, Yoshida, M. | | Deposit date: | 2010-10-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.544 Å) | | Cite: | Discovery and biological activity of a novel class I PI3K inhibitor, CH5132799
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4K7D
 
 | | Crystal Structure of Parkin C-terminal RING domains | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, MALONATE ION, ... | | Authors: | Sauve, V, Trempe, J.-F, Menade, M, Gehring, K. | | Deposit date: | 2013-04-17 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of parkin reveals mechanisms for ubiquitin ligase activation.
Science, 340, 2013
|
|
3RZF
 
 | | Crystal Structure of Inhibitor of kappaB kinase beta (I4122) | | Descriptor: | (4-{[4-(4-chlorophenyl)pyrimidin-2-yl]amino}phenyl)[4-(2-hydroxyethyl)piperazin-1-yl]methanone, MGC80376 protein | | Authors: | Xu, G, Lo, Y.C, Li, Q, Napolitano, G, Wu, X, Jiang, X, Dreano, M, Karin, M, Wu, H. | | Deposit date: | 2011-05-11 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structure of inhibitor of KappaB kinase Beta.
Nature, 472, 2011
|
|
3S0Y
 
 | |
3ANX
 
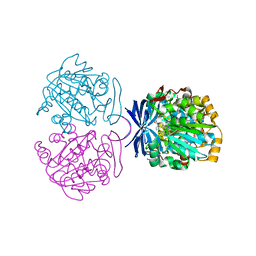 | | Crystal structure of triamine/agmatine aminopropyltransferase (SPEE) from thermus thermophilus, complexed with MTA | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, spermidine synthase | | Authors: | Ganbe, T, Ohnuma, M, Sato, T, Tanaka, N, Oshima, T, Kumasaka, T. | | Deposit date: | 2010-09-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures and enzymatic properties of a triamine/agmatine aminopropyltransferase from Thermus thermophilus
J.Mol.Biol., 408, 2011
|
|
1KBQ
 
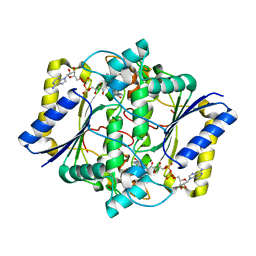 | | Complex of Human NAD(P)H quinone Oxidoreductase with 5-methoxy-1,2-dimethyl-3-(4-nitrophenoxymethyl)indole-4,7-dione (ES936) | | Descriptor: | 5-METHOXY-1,2-DIMETHYL-3-(4-NITROPHENOXYMETHYL)INDOLE-4,7-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Faig, M, Bianchet, M.A, Amzel, L.M. | | Deposit date: | 2001-11-06 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a mechanism-based inhibitor of NAD(P)H:quinone oxidoreductase 1 by biochemical, X-ray crystallographic, and mass spectrometric approaches.
Biochemistry, 40, 2001
|
|
3WLI
 
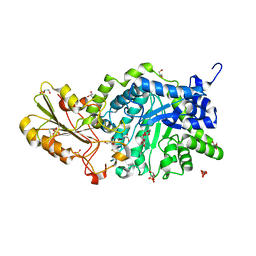 | | Crystal Structure Analysis of Plant Exohydrolase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, GLYCEROL, ... | | Authors: | Streltsov, V.A, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
