2AWG
 
 | | Structure of the PPIase domain of the Human FK506-binding protein 8 | | Descriptor: | 38 kDa FK-506 binding protein | | Authors: | Walker, J.R, Davis, T, Newman, E.M, Finerty, P, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-09-01 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the human FK-506 binding protein 8
To be Published
|
|
2AWW
 
 | | Synapse associated protein 97 PDZ2 domain variant C378G with C-terminal GluR-A peptide | | Descriptor: | 18-residue C-terminal peptide from glutamate receptor, ionotropic, AMPA1, ... | | Authors: | Von Ossowski, I, Oksanen, E, Von Ossowski, L, Cai, C, Sundberg, M, Goldman, A, Keinanen, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of the second PDZ domain of SAP97 in complex with a GluR-A C-terminal peptide
Febs J., 273, 2006
|
|
2AP6
 
 | | X-Ray Crystal Structure of Protein Atu4242 from Agrobacterium tumefaciens. Northeast Strucutral Genomics Consortium Target AtR43. | | Descriptor: | hypothetical protein Atu4242 | | Authors: | Benach, J, Kuzin, A.P, Forouhar, F, Abashidze, M, Vorobiev, S.M, Rong, X, Acton, T.B, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a Hypothetical protein Atu4242 from Agrobacterium tumefaciens (strain C58 / ATCC 3 NESG Target ATR43.
To be Published
|
|
6LXB
 
 | | X-ray structure of human PPARalpha ligand binding domain-saroglitazar co-crystals obtained by soaking | | Descriptor: | (2S)-2-ethoxy-3-[4-[2-[2-methyl-5-(4-methylsulfanylphenyl)pyrrol-1-yl]ethoxy]phenyl]propanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Honda, A, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
2AS8
 
 | | Crystal structure of mature and fully active Der p 1 allergen | | Descriptor: | MAGNESIUM ION, Major mite fecal allergen Der p 1 | | Authors: | de Halleux, S, Stura, E, VanderElst, L, Carlier, V, Jacquemin, M, Saint-Remy, J.-M. | | Deposit date: | 2005-08-23 | | Release date: | 2005-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-dimensional structure and IgE-binding properties of mature fully active Der p 1, a clinically relevant major allergen
J.Allergy Clin.Immunol., 117, 2006
|
|
9ATW
 
 | | Structure of biofilm-forming functional amyloid PSMa1 from Staphylococcus aureus | | Descriptor: | Phenol-soluble modulin alpha 1 peptide | | Authors: | Hansen, K.H, Byeon, C.H, Liu, Q, Drace, T, Boesen, T, Conway, J.F, Andreasen, M, Akbey, U. | | Deposit date: | 2024-02-27 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of biofilm-forming functional amyloid PSM alpha 1 from Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8Y6V
 
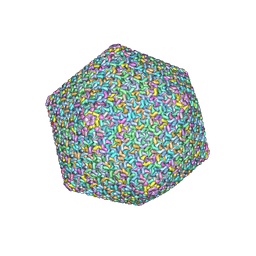 | | Near-atomic structure of icosahedrally averaged jumbo bacteriophage PhiKZ capsid | | Descriptor: | gp119, gp120, gp162, ... | | Authors: | Yang, Y, Shao, Q, Guo, M, Han, L, Zhao, X, Wang, A, Li, X, Wang, B, Pan, J, Chen, Z, Fokine, A, Sun, L, Fang, Q. | | Deposit date: | 2024-02-03 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Capsid structure of bacteriophage Phi KZ provides insights into assembly and stabilization of jumbo phages.
Nat Commun, 15, 2024
|
|
2AW5
 
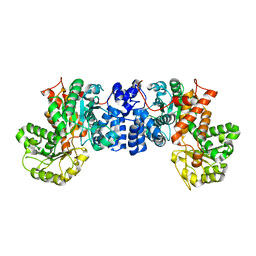 | | Crystal structure of a human malic enzyme | | Descriptor: | NADP-dependent malic enzyme | | Authors: | Papagrigoriou, E, Berridge, G, Smee, C, Bray, J, Arrowsmith, C, Edwards, A, Weigelt, J, Sundstrom, M, Oppermann, U, Gileadi, O, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-31 | | Release date: | 2005-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a human malic enzyme
To be published
|
|
2AXO
 
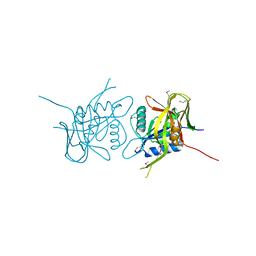 | | X-Ray Crystal Structure of Protein AGR_C_4864 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR35. | | Descriptor: | hypothetical protein Atu2684 | | Authors: | Forouhar, F, Abashidze, M, Benach, J, Xiao, R, Janjua, H, Conover, K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-09-05 | | Release date: | 2005-09-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Hypothetical Protein AGR_C_4864 from Agrobacterium tumefaciens, NESG target AtR35
To be Published
|
|
2AYI
 
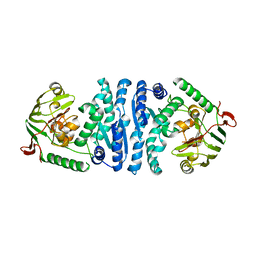 | | Wild-type AmpT from Thermus thermophilus | | Descriptor: | Aminopeptidase T, ZINC ION | | Authors: | Odintsov, S.G, Sabala, I, Bourenkov, G, Rybin, V, Bochtler, M. | | Deposit date: | 2005-09-07 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Substrate Access to the Active Sites in Aminopeptidase T, a Representative of a New Metallopeptidase Clan.
J.Mol.Biol., 354, 2005
|
|
7Y8M
 
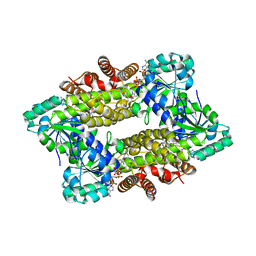 | | Structure of ScIRED-R2-V3 from Streptomyces clavuligerus in complex with 5-(3-fluorophenyl)-3,4-dihydro-2H-pyrrole | | Descriptor: | 2-[2,5-bis(fluoranyl)phenyl]pyrrolidine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, reductase | | Authors: | Zhang, L.L, Liu, W.D, Shi, M, Huang, J.W, Yang, Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Engineered Imine Reductase for Larotrectinib Intermediate Manufacture
Acs Catalysis, 12, 2022
|
|
5OVK
 
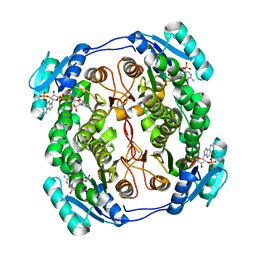 | | Crystal structure MabA bound to NADPH from M. smegmatis | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kussau, T, Van Wyk, N, Viljoen, A, Olieric, V, Flipo, M, Kremer, L, Blaise, M. | | Deposit date: | 2017-08-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural rearrangements occurring upon cofactor binding in the Mycobacterium smegmatis beta-ketoacyl-acyl carrier protein reductase MabA.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
2AZM
 
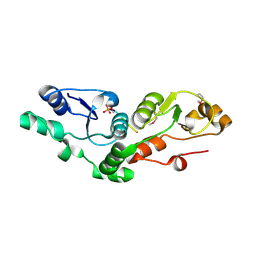 | | Crystal structure of the MDC1 brct repeat in complex with the histone tail of gamma-H2AX | | Descriptor: | GAMMA-H2AX HISTONE, Mediator of DNA damage checkpoint protein 1 | | Authors: | Clapperton, J.A, Stucki, M, Mohammad, D, Yaffe, M.B, Jackson, S.P, Smerdon, S.J. | | Deposit date: | 2005-09-12 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | MDC1 Directly Binds Phosphorylated Histone H2AX to Regulate Cellular Responses to DNA Double-Strand Breaks
Cell(Cambridge,Mass.), 123, 2005
|
|
6LLA
 
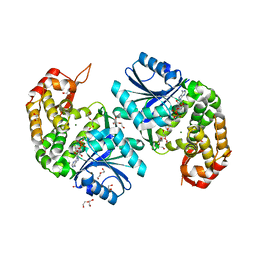 | | Crystal structure of Providencia alcalifaciens 3-dehydroquinate synthase (DHQS) in complex with Mg2+ and NAD | | Descriptor: | 1,2-ETHANEDIOL, 3-dehydroquinate synthase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Neetu, N, Katiki, M, Kumar, P. | | Deposit date: | 2019-12-22 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and Biochemical Analyses Reveal that Chlorogenic Acid Inhibits the Shikimate Pathway.
J.Bacteriol., 202, 2020
|
|
8ZFJ
 
 | | cryo-EM structure of GPR4-Gs complex at pH 8.5 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Ma, Y, Tang, M, Ru, H, Song, G. | | Deposit date: | 2024-05-07 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | cryo-EM structure of GPR4-Gs complex at pH 8.5
to be published
|
|
2AQN
 
 | | CU/ZN superoxide dismutase from neisseria meningitidis | | Descriptor: | COPPER (I) ION, COPPER (II) ION, SULFATE ION, ... | | Authors: | DiDonato, M, Kassmann, C.J, Bruns, C.K, Cabelli, D.E, Cao, Z, Tabatabai, L.B, Kroll, J.S, Getzoff, E.D. | | Deposit date: | 2005-08-18 | | Release date: | 2006-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | CU/ZN superoxide dismutase from neisseria meningitidis
To be Published
|
|
2ARV
 
 | | Structure of human Activin A | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, GLYCEROL, Inhibin beta A chain, ... | | Authors: | Harrington, A.E, Morris-Triggs, S.A, Ruotolo, B.T, Robinson, C.V, Ohnuma, S, Hyvonen, M. | | Deposit date: | 2005-08-22 | | Release date: | 2006-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the inhibition of activin signalling by follistatin
Embo J., 25, 2006
|
|
2AHK
 
 | | Crystal structure of the met-form of the copper-bound Streptomyces castaneoglobisporus tyrosinase in complex with a caddie protein obtained by soking in cupric sulfate for 6 months | | Descriptor: | CADDIE PROTEIN ORF378, COPPER (II) ION, NITRATE ION, ... | | Authors: | Matoba, Y, Kumagai, T, Yamamoto, A, Yoshitsu, H, Sugiyama, M. | | Deposit date: | 2005-07-28 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystallographic Evidence That the Dinuclear Copper Center of Tyrosinase Is Flexible during Catalysis
J.Biol.Chem., 281, 2006
|
|
2AV6
 
 | |
2AWE
 
 | |
2ALX
 
 | | Ribonucleotide Reductase R2 from Escherichia coli in space group P6(1)22 | | Descriptor: | MANGANESE (II) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 | | Authors: | Sommerhalter, M, Saleh, L, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2005-08-08 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Escherichia coli ribonucleotide reductase R2 in space group P6122.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
8ZUG
 
 | |
8ZMF
 
 | | Crystal structure of an inverse agonist antipsychotic drug derivative-bound 5-HT2C | | Descriptor: | 1-[(4-fluorophenyl)methyl]-1-[(8~{S})-5-methyl-5-azaspiro[2.5]octan-8-yl]-3-[[4-(2-methylpropoxy)phenyl]methyl]urea, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562 | | Authors: | Oguma, T, Asada, H, Sekiguchi, Y, Imono, M, Iwata, S, Kusakabe, K. | | Deposit date: | 2024-05-23 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Dual 5-HT 2A and 5-HT 2C Receptor Inverse Agonist That Affords In Vivo Antipsychotic Efficacy with Minimal hERG Inhibition for the Treatment of Dementia-Related Psychosis.
J.Med.Chem., 67, 2024
|
|
2AZV
 
 | | Solution structure of the T22G mutant of N-terminal SH3 domain of DRK (calculated without NOEs) | | Descriptor: | SH2-SH3 adapter protein drk | | Authors: | Bezsonova, I, Singer, A.U, Choy, W.-Y, Tollinger, M, Forman-Kay, J.D. | | Deposit date: | 2005-09-12 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Comparison of the Unstable drkN SH3 Domain and a Stable Mutant
Biochemistry, 44, 2005
|
|
2B1F
 
 | | Antiparallel four-stranded coiled coil specified by a 3-3-1 hydrophobic heptad repeat | | Descriptor: | General control protein GCN4 | | Authors: | Deng, Y, Liu, J, Zheng, Q, Eliezer, D, Kallenbach, N.R, Lu, M. | | Deposit date: | 2005-09-15 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Antiparallel four-stranded coiled coil specified by a 3-3-1 hydrophobic heptad repeat.
Structure, 14, 2006
|
|
