6YVL
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 1.42 MGy exposure | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YVM
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 2.13 MGy exposure | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6E57
 
 | | Bacteroides ovatus mixed-linkage glucan utilization locus (MLGUL) SGBP-B in complex with mixed-linkage heptasaccharide | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Koropatkin, N.M, Schnizlein, M, Bahr, C.M.E. | | Deposit date: | 2018-07-19 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Surface glycan-binding proteins are essential for cereal beta-glucan utilization by the human gut symbiont Bacteroides ovatus.
Cell.Mol.Life Sci., 76, 2019
|
|
6E5V
 
 | | human mGlu8 receptor amino terminal domain in complex with (S)-3,4-Dicarboxyphenylglycine (DCPG) | | Descriptor: | 4-[(S)-amino(carboxy)methyl]benzene-1,2-dicarboxylic acid, CHLORIDE ION, Metabotropic glutamate receptor 8 | | Authors: | Chen, Q, Ho, J.D, Ashok, S, Vargas, M.C, Wang, J, Atwell, S, Bures, M, Schkeryantz, J.M, Monn, J.A, Hao, J. | | Deposit date: | 2018-07-23 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Basis for ( S)-3,4-Dicarboxyphenylglycine (DCPG) As a Potent and Subtype Selective Agonist of the mGlu8Receptor.
J. Med. Chem., 61, 2018
|
|
6YVN
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 2.84 MGy exposure | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6EOK
 
 | | Crystal structure of E. coli L-asparaginase II | | Descriptor: | L-asparaginase 2, ZINC ION | | Authors: | Cerofolini, L, Giuntini, S, Carlon, A, Ravera, E, Calderone, V, Fragai, M, Parigi, G, Luchinat, C. | | Deposit date: | 2017-10-09 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of PEGylated Asparaginase: New Opportunities from NMR Analysis of Large PEGylated Therapeutics.
Chemistry, 25, 2019
|
|
8DR0
 
 | | Closed state of RFC:PCNA bound to a 3' ss/dsDNA junction | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*GP*GP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*CP*GP*GP*GP*GP*GP*GP*GP*CP*CP*CP*CP*GP*GP*GP*G)-3'), GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8EBC
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria monocytogenes in the complex with IMP | | Descriptor: | FORMIC ACID, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Osipiuk, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-08-31 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria monocytogenes in the complex with IMP
To Be Published
|
|
6E8F
 
 | |
6YWU
 
 | | Human OMPD-domain of UMPS (K314AcK) in complex with UMP at 1.1 Angstroms resolution | | Descriptor: | GLYCEROL, SULFATE ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-30 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YWT
 
 | |
6YVO
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 3.55 MGy exposure | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-28 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
8DQX
 
 | | Open state of RFC:PCNA bound to a 3' ss/dsDNA junction | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*T)-3'), DNA (5'-D(P*TP*CP*CP*GP*AP*GP*CP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*GP*CP*CP*CP*GP*GP*A)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
6EB5
 
 | |
6E85
 
 | | 1.25 Angstrom Resolution Crystal Structure of 4-hydroxythreonine-4-phosphate Dehydrogenase from Klebsiella pneumoniae. | | Descriptor: | CHLORIDE ION, D-threonate 4-phosphate dehydrogenase, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-27 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6E8Q
 
 | | S. CEREVISIAE CYP51 COMPLEXED WITH Posaconazole | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Lanosterol 14-alpha demethylase, PENTAETHYLENE GLYCOL, ... | | Authors: | Tyndall, J.D, Keniya, M.V, Sabherwal, M, Monk, B.C. | | Deposit date: | 2018-07-30 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Azole Resistance Reduces Susceptibility to the Tetrazole Antifungal VT-1161.
Antimicrob. Agents Chemother., 63, 2019
|
|
8DR6
 
 | | Closed state of RFC:PCNA bound to a nicked dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (32-MER), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DQZ
 
 | | Intermediate state of RFC:PCNA bound to a 3' ss/dsDNA junction | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*GP*GP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*CP*GP*GP*GP*GP*GP*GP*GP*CP*CP*CP*CP*GP*GP*GP*G)-3'), GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
7XLJ
 
 | | The crystal structure of ORE1(ANAC092) NAC domain | | Descriptor: | NAC domain-containing protein 92 | | Authors: | Chun, I.S, Kim, M.S. | | Deposit date: | 2022-04-21 | | Release date: | 2023-03-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of DNA binding by the NAC transcription factor ORE1, a master regulator of plant senescence.
Plant Commun., 4, 2023
|
|
5OML
 
 | | Crystal structure of Trypanosoma Brucei PEX14 N-terminal domain in complex with small molecules to investigate the water envelope | | Descriptor: | (3~{R})-3-[[1-(2-hydroxyethyl)-5-[(4-methoxynaphthalen-1-yl)methyl]-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-3-yl]carbonylamino]-3-phenyl-propanoic acid, BETA-MERCAPTOETHANOL, Peroxin 14, ... | | Authors: | Ratkova, E.L, Dawidowski, M, Napolitano, V, Dubin, G, Fino, R, Popowicz, G, Sattler, M, Tetko, I.V. | | Deposit date: | 2017-08-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Trypanosoma Brucei PEX14 N-terminal domain in complex with small molecules to investigate the water envelope
To Be Published
|
|
6ZQW
 
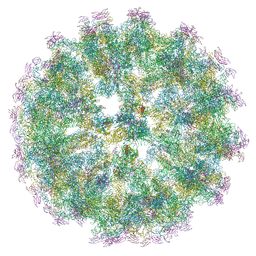 | | Cryo-EM structure of immature Spondweni virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, prM | | Authors: | Renner, M, Dejnirattisai, W, Carrique, L, Serna Martin, I, Karia, D, Ilca, S.L, Ho, S.F, Kotecha, A, Keown, J.R, Mongkolsapaya, J, Screaton, G.R, Grimes, J.M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Flavivirus maturation leads to the formation of an occupied lipid pocket in the surface glycoproteins.
Nat Commun, 12, 2021
|
|
6ECJ
 
 | |
6ZRV
 
 | | Crystal Structure of Stabilized Active Plasminogen Activator Inhibitor-1 (PAI-1-W175F) in Complex with an Inhibitory Nanobody (VHH-s-a93, Nb93) | | Descriptor: | Plasminogen activator inhibitor 1, VHH-s-a93 (Nb93) | | Authors: | Sillen, M, Weeks, S.D, Strelkov, S.V, Declerck, P.J. | | Deposit date: | 2020-07-15 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Insights into the Mechanism of a Nanobody That Stabilizes PAI-1 and Modulates Its Activity.
Int J Mol Sci, 21, 2020
|
|
5OSX
 
 | | Translation initiation factor 4E in complex with m7G(5'S)ppp(5'S)G mRNA 5' cap analog | | Descriptor: | Eukaryotic translation initiation factor 4E, POTASSIUM ION, [(2~{S},3~{S},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-3~{H}-purin-7-ium-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl-[[[(3~{R},4~{S})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-phosphinic acid | | Authors: | Warminski, M, Nowak, E, Wojtczak, B.A, Fac-Dabrowska, K, Kubacka, D, Nowicka, A, Sikorski, P.J, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2017-08-18 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | 5'-Phosphorothiolate Dinucleotide Cap Analogues: Reagents for Messenger RNA Modification and Potent Small-Molecular Inhibitors of Decapping Enzymes.
J. Am. Chem. Soc., 140, 2018
|
|
6ZS3
 
 | | Crystal structure of the fifth bromodomain of human protein polybromo-1 in complex with 2-(6-amino-5-(piperazin-1-yl)pyridazin-3-yl)phenol | | Descriptor: | 1,2-ETHANEDIOL, 2-(6-azanyl-5-piperazin-4-ium-1-yl-pyridazin-3-yl)phenol, Protein polybromo-1 | | Authors: | Preuss, F, Joerger, A.C, Wanior, M, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-07-15 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Pan-SMARCA/PB1 Bromodomain Inhibitors and Their Role in Regulating Adipogenesis.
J.Med.Chem., 63, 2020
|
|
