6Z98
 
 | | NMR solution structure of the peach allergen Pru p 1.0101 | | Descriptor: | Major allergen Pru p 1 | | Authors: | Eidelpes, R, Fuehrer, S, Hofer, F, Kamenik, A.S, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Zeatin Binding of the Peach Allergen Pru p 1 .
J.Agric.Food Chem., 69, 2021
|
|
6R73
 
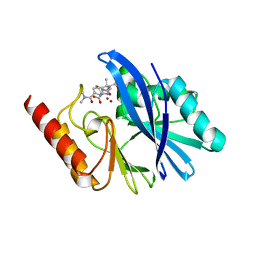 | | Structure of IMP-13 metallo-beta-lactamase complexed with hydrolysed meropenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Beta-lactamase, ZINC ION | | Authors: | Softley, C.A, Zak, K, Kolonko, M, Sattler, M, Popowicz, G. | | Deposit date: | 2019-03-28 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
5FP6
 
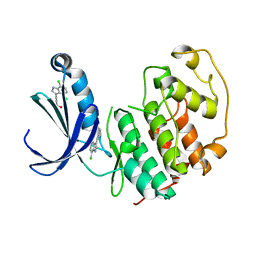 | | Structure of cyclin-dependent kinase 2 with small-molecule ligand 3-(4,7-dichloro-1H-indol-3-yl)prop-2-yn-1-ol (AT17833) in an alternate binding site. | | Descriptor: | 3-(4,7-dichloro-1H-indol-3-yl)prop-2-yn-1-ol, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Jhoti, H, Ludlow, R.F, O'Reilly, M, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-11-27 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6R78
 
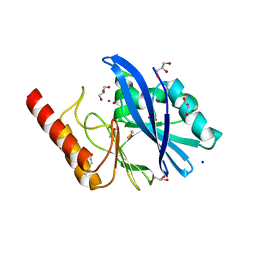 | | Structure of IMP-13 metallo-beta-lactamase in apo form (loop closed) | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Beta-lactamase, ... | | Authors: | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | Deposit date: | 2019-03-28 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
6R7V
 
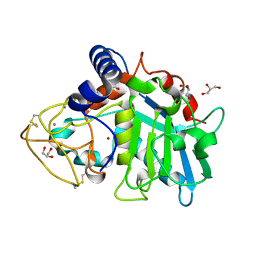 | | Tannerella forsythia promirolysin mutant E225A | | Descriptor: | CALCIUM ION, GLYCEROL, Mirolysin, ... | | Authors: | Rodriguez-Banqueri, A, Guevara, T, Ksiazek, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2019-03-29 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based mechanism of cysteine-switch latency and of catalysis by pappalysin-family metallopeptidases.
Iucrj, 7, 2020
|
|
2P2Z
 
 | | Crystal structure of TTHB049 from Thermus thermophilus HB8 | | Descriptor: | Alpha-ribazole-5'-phosphate phosphatase, GLYCEROL, SODIUM ION | | Authors: | Sugahara, M, Taketa, M, Tanaka, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-08 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of TTHB049 from Thermus thermophilus HB8
To be Published
|
|
6ZBK
 
 | | Crystal structure of the human complex between RPAP3 and TRBP | | Descriptor: | RISC-loading complex subunit TARBP2, RNA polymerase II-associated protein 3 | | Authors: | Charron, C, Abel, Y, Charpentier, B, Rederstorff, M. | | Deposit date: | 2020-06-08 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The interaction between RPAP3 and TRBP reveals a possible involvement of the HSP90/R2TP chaperone complex in the regulation of miRNA activity.
Nucleic Acids Res., 50, 2022
|
|
6ZE1
 
 | | human NBD1 of CFTR in complex with nanobody G11a | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, ... | | Authors: | Garcia-Pino, A, Govaerts, C, Scholl, D, Sigoillot, M. | | Deposit date: | 2020-06-15 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | A topological switch in CFTR modulates channel activity and sensitivity to unfolding.
Nat.Chem.Biol., 17, 2021
|
|
5C8E
 
 | |
5PB7
 
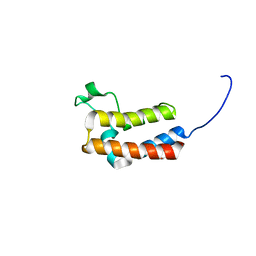 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B in complex with N09440a | | Descriptor: | 1,2-ETHANEDIOL, 1~{H}-indazol-5-amine, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4HS8
 
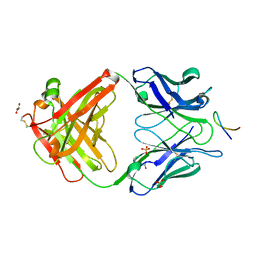 | |
5PBG
 
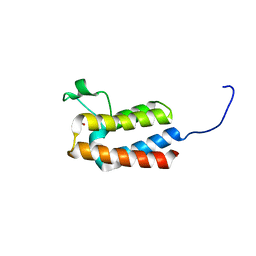 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 1) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PBT
 
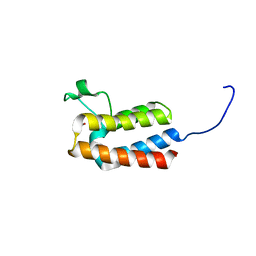 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 14) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PCA
 
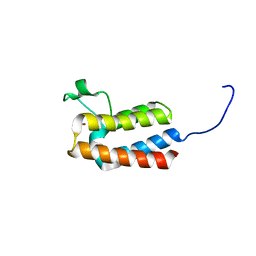 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 31) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
7PIG
 
 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN088 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-azanyl-2-chloranyl-3-[(Z)-piperidin-3-ylidenemethyl]benzenecarbonitrile, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
5U6I
 
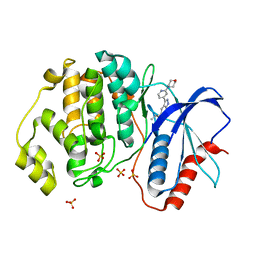 | | Discovery of MLi-2, an Orally Available and Selective LRRK2 Inhibitor that Reduces Brain Kinase Activity | | Descriptor: | 3-[2-(morpholin-4-yl)pyridin-4-yl]-5-[(propan-2-yl)oxy]-1H-indazole, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Scott, J.D, DeMong, D.E, Fell, M.J, Mirescu, C, Basu, K, Greshock, T.J, Morrow, J.A, Xiao, L, Hruza, A, Harris, J, Tiscia, H.E, Chang, R.K, Embrey, M.W, McCauley, J.A, Li, W, Lin, S, Liu, H, Dai, X, Baptista, M, Agnihotri, G, Columbus, J, Mei, H, Poirier, M, Zhou, X, Lin, Y, Yin, Z, Sanders, J.M, Drolet, R.E, Kern, J.T, Kennedy, M.E, Parker, E.M, Stamford, A.W, Nargund, R, Miller, M.W. | | Deposit date: | 2016-12-08 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of a 3-(4-Pyrimidinyl) Indazole (MLi-2), an Orally Available and Selective Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitor that Reduces Brain Kinase Activity.
J. Med. Chem., 60, 2017
|
|
6BCC
 
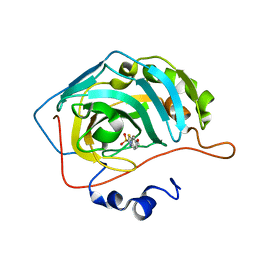 | | Joint X-ray/neutron structure of human carbonic anhydrase II in complex with ethoxzolamide | | Descriptor: | 6-ethoxy-1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Kovalevsky, A, McKenna, R, Aggarwal, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | "To Be or Not to Be" Protonated: Atomic Details of Human Carbonic Anhydrase-Clinical Drug Complexes by Neutron Crystallography and Simulation.
Structure, 26, 2018
|
|
5PCK
 
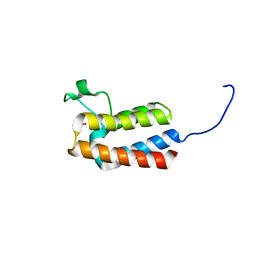 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 41) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PCX
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 54) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
7PIE
 
 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN068 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-azanyl-5-[3-[(2R)-morpholin-2-yl]phenyl]benzenecarbonitrile, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.427 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
4LQ3
 
 | | Crystal structure of human norovirus RNA-dependent RNA-polymerase bound to the inhibitor PPNDS | | Descriptor: | 3-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]-7-nitronaphthalene-1,5-disulfonic acid, 5'-R(P*GP*G)-3', MAGNESIUM ION, ... | | Authors: | Milani, M, Tarantino, D, Mastrangelo, E, Croci, R. | | Deposit date: | 2013-07-17 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Naphthalene-sulfonate inhibitors of human norovirus RNA-dependent RNA-polymerase.
Antiviral Res., 102, 2014
|
|
7PID
 
 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN060 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-azanyl-5-[3-[(3R)-morpholin-3-yl]phenyl]benzenecarbonitrile, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
5PBK
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 5) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PDE
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 70) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
7PIF
 
 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN086 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-[3-[4-(aminomethyl)oxan-4-yl]phenyl]-2-azanyl-benzenecarbonitrile, CHLORIDE ION, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
