4XM4
 
 | |
4U0S
 
 | | Structure of Eukaryotic fic domain containing protein with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, MAGNESIUM ION, ... | | Authors: | Cole, A.R, Katan, M, Bunney, T.D. | | Deposit date: | 2014-07-14 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of the human, FIC-domain containing protein HYPE and implications for its functions.
Structure, 22, 2014
|
|
4TUO
 
 | | Crystal structure of monoclonal antibody against neuroblastoma associated antigen. | | Descriptor: | Heavy chain of monoclonal antibody against neuroblastoma associated antigen, Light chain of monoclonal antibody against neuroblastoma associated antigen, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-3)-[2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)]beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Golik, P, Grudnik, P, Horwacik, I, Zdzalik, M, Rokita, H, Dubin, G. | | Deposit date: | 2014-06-24 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis of GD2 Ganglioside and Mimetic Peptide Recognition by 14G2a Antibody.
Mol.Cell Proteomics, 14, 2015
|
|
6YJF
 
 | | Plasmoodium vivax phosphoglycerate kinase bound to nitrofuran inhibitor from PEGSmear at pH 6.5 | | Descriptor: | (2~{S})-2-(5-nitrofuran-2-yl)-2,3,5,6,7,8-hexahydro-1~{H}-[1]benzothiolo[2,3-d]pyrimidin-4-one, GLYCEROL, Phosphoglycerate kinase | | Authors: | Hyvonen, M, Brear, P, Blaszczyk, B.K. | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Phosphoglycerate Kinase as a potential target for antimalarial therapy
to be published
|
|
7ZPJ
 
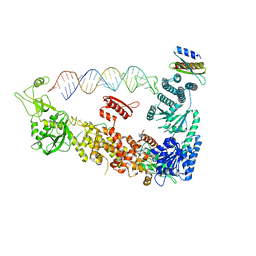 | | Mammalian Dicer in the "pre-dicing state" with pre-miR-15a substrate and TARBP2 subunit | | Descriptor: | 59-nt precursor of miR-15a, Endoribonuclease Dicer, RISC-loading complex subunit TARBP2 isoform 1 | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-04-27 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
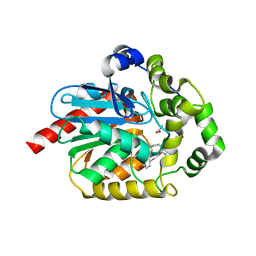
6YN2
 
 | |
7ZKY
 
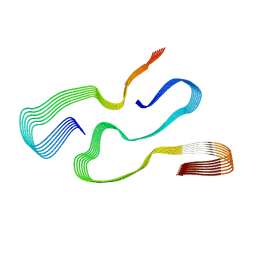 | |
7ZYV
 
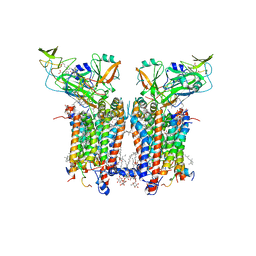 | | Cryo-EM structure of catalytically active Spinacia oleracea cytochrome b6f in complex with endogenous plastoquinones at 2.13 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, BETA-CAROTENE, ... | | Authors: | Sarewicz, M, Szwalec, M, Pintscher, S, Indyka, P, Rawski, M, Pietras, R, Mielecki, B, Koziej, L, Jaciuk, M, Glatt, S, Osyczka, A. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | High-resolution cryo-EM structures of plant cytochrome b 6 f at work.
Sci Adv, 9, 2023
|
|
3MUJ
 
 | | Early B-cell factor 3 (EBF3) IPT/TIG and dimerization helices | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Transcription factor COE3 | | Authors: | Siponen, M.I, Lehtio, L, Arrowsmith, C.H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schueler, H, Schutz, P, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Berglund, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Determination of Functional Domains in Early B-cell Factor (EBF) Family of Transcription Factors Reveals Similarities to Rel DNA-binding Proteins and a Novel Dimerization Motif.
J.Biol.Chem., 285, 2010
|
|
7OTC
 
 | | Cryo-EM structure of an Escherichia coli 70S ribosome in complex with elongation factor G and the antibiotic Argyrin B | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Wieland, M, Koller, T.O, Wilson, D.N. | | Deposit date: | 2021-06-10 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The cyclic octapeptide antibiotic argyrin B inhibits translation by trapping EF-G on the ribosome during translocation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4UF1
 
 | | Deerpox virus DPV022 in complex with Bak BH3 | | Descriptor: | Antiapoptotic membrane protein, Bcl-2 homologous antagonist/killer, SULFATE ION | | Authors: | Burton, D.R, Kvansakul, M. | | Deposit date: | 2014-12-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Deerpox Virus-Mediated Inhibition of Apoptosis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7P9O
 
 | | Structure of E.coli RlmJ in complex with a SAM analogue (CA) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]amino}-5'-deoxyadenosine, Ribosomal RNA large subunit methyltransferase J | | Authors: | Meynier, V, Catala, M, Oerum, S, Barraud, P, Tisne, C. | | Deposit date: | 2021-07-27 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Synthesis of RNA-cofactor conjugates and structural exploration of RNA recognition by an m6A RNA methyltransferase.
Nucleic Acids Res., 50, 2022
|
|
4UOW
 
 | | Crystal structure of the titin M10-Obscurin Ig domain 1 complex | | Descriptor: | CHLORIDE ION, Obscurin, SODIUM ION, ... | | Authors: | Pernigo, S, Fukuzawa, A, Gautel, M, Steiner, R.A. | | Deposit date: | 2014-06-10 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of the Human Titin:Obscurin Complex Reveals a Conserved Yet Specific Muscle M-Band Zipper Module.
J.Mol.Biol., 427, 2015
|
|
1C0K
 
 | | CRYSTAL STRUCTURE ANALYSIS OF D-AMINO ACID OXIDASE IN COMPLEX WITH L-LACTATE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LACTIC ACID, PROTEIN (D-AMINO ACID OXIDASE) | | Authors: | Umhau, S, Molla, G, Diederichs, K, Pilone, M.S, Ghisla, S, Welte, W, Pollegioni, L. | | Deposit date: | 1999-07-16 | | Release date: | 2000-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The x-ray structure of D-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
7BWI
 
 | | Solution structure of recombinant APETx1 | | Descriptor: | Kappa-actitoxin-Ael2a | | Authors: | Matsumura, K, Kobayashi, N, Kurita, J, Nishimura, Y, Yokogawa, M, Imai, S, Shimada, I, Osawa, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Mechanism of hERG inhibition by gating-modifier toxin, APETx1, deduced by functional characterization.
Bmc Mol Cell Biol, 22, 2021
|
|
1C3R
 
 | |
6XKP
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CV07-270 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV07-270 Heavy Chain, CV07-270 Light Chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.
Cell, 183, 2020
|
|
1BP7
 
 | | GROUP I MOBILE INTRON ENDONUCLEASE I-CREI COMPLEXED WITH HOMING SITE DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*TP* GP*C)-3'), DNA (5'-D(*GP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*TP* CP*G)-3'), ... | | Authors: | Jurica, M.S, Monnat Junior, R.J, Stoddard, B.L. | | Deposit date: | 1998-08-13 | | Release date: | 1999-01-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-CreI.
Mol.Cell, 2, 1998
|
|
7QDN
 
 | |
4UMX
 
 | | IDH1 R132H in complex with cpd 1 | | Descriptor: | 2,6-bis(1H-imidazol-1-ylmethyl)-4-(2,4,4-trimethylpentan-2-yl)phenol, GLYCEROL, ISOCITRATE DEHYDROGENASE [NADP] CYTOPLASMIC, ... | | Authors: | Mathieu, M, Marquette, J.P. | | Deposit date: | 2014-05-22 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Selective Inhibition of Mutant Isocitrate Dehydrogenase 1 (Idh1) Via Disruption of a Metal Binding Network by an Allosteric Small Molecule.
J.Biol.Chem., 290, 2015
|
|
8R5J
 
 | | Crystal structure of MERS-CoV main protease | | Descriptor: | Non-structural protein 11 | | Authors: | Balcomb, B.H, Fairhead, M, Koekemoer, L, Lithgo, R.M, Aschenbrenner, J.C, Chandran, A.V, Godoy, A.S, Lukacik, P, Marples, P.G, Mazzorana, M, Ni, X, Strain-Damerell, C, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-11-16 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal structure of MERS-CoV main protease
To Be Published
|
|
7QCO
 
 | | The structure of Photosystem I tetramer from Chroococcidiopsis TS-821, a thermophilic, unicellular, non-heterocyst-forming cyanobacterium | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Semchonok, D.A, Mondal, J, Cooper, J.C, Schlum, K, Li, M, Amin, M, Sorzano, C.O.S, Ramirez-Aportela, E, Kastritis, P.L, Boekema, E.J, Guskov, A, Bruce, B.D. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a tetrameric photosystem I from Chroococcidiopsis TS-821, a thermophilic, unicellular, non-heterocyst-forming cyanobacterium.
Plant Commun., 3, 2022
|
|
7QK4
 
 | | EED in complex with PRC2 allosteric inhibitor compound 22 (MAK683) | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-[(5-fluoranyl-2,3-dihydro-1-benzofuran-4-yl)methyl]-8-(2-methylpyridin-3-yl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
7QJU
 
 | | EED in complex with PRC2 allosteric inhibitor compound 7 | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-[4-[(dimethylamino)methyl]phenyl]-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
6YEL
 
 | | Stromal interaction molecule 1 coiled-coil 1 fragment | | Descriptor: | Stromal interaction molecule 1 | | Authors: | Rathner, P, Cerofolini, L, Ravera, E, Bechmann, M, Grabmayr, H, Fahrner, M, Fragai, M, Romanin, C, Luchinat, C, Mueller, N. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-02 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Interhelical interactions within the STIM1 CC1 domain modulate CRAC channel activation.
Nat.Chem.Biol., 17, 2021
|
|
