8ZND
 
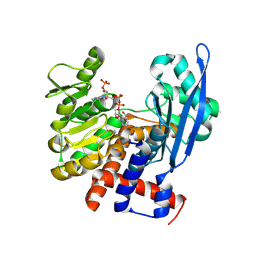 | |
3ZL7
 
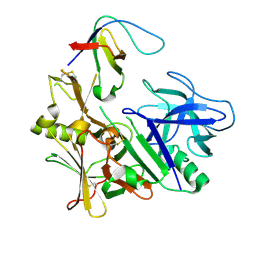 | | BACE2 FYNOMER COMPLEX | | Descriptor: | BETA-SECRETASE 2, FYNOMER 2B-H11 | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-28 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1HMK
 
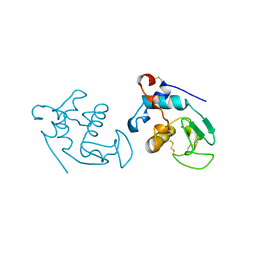 | | RECOMBINANT GOAT ALPHA-LACTALBUMIN | | Descriptor: | CALCIUM ION, PROTEIN (ALPHA-LACTALBUMIN) | | Authors: | Horii, K, Matsushima, M, Tsumoto, K, Kumagai, I. | | Deposit date: | 1998-11-26 | | Release date: | 1999-11-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effect of the extra n-terminal methionine residue on the stability and folding of recombinant alpha-lactalbumin expressed in Escherichia coli.
J.Mol.Biol., 285, 1999
|
|
3ZMQ
 
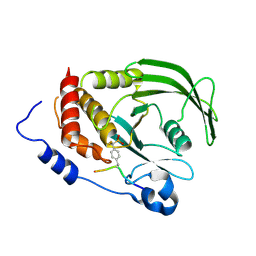 | | Src-derived mutant peptide inhibitor complex of PTP1B | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Temmerman, K, Pogenberg, V, Meyer, C, Koehn, M, Wilmanns, M. | | Deposit date: | 2013-02-12 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Development of Accessible Peptidic Tool Compounds to Study the Phosphatase Ptp1B in Intact Cells.
Acs Chem.Biol., 9, 2014
|
|
3ZPH
 
 | |
3BXU
 
 | | PpcB, A Cytochrome c7 from Geobacter sulfurreducens | | Descriptor: | Cytochrome c3, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2008-01-14 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into the modulation of the redox properties of two Geobacter sulfurreducens homologous triheme cytochromes.
Biochim.Biophys.Acta, 1777, 2008
|
|
3ZJP
 
 | | M.acetivorans protoglobin in complex with imidazole | | Descriptor: | GLYCEROL, IMIDAZOLE, PHOSPHATE ION, ... | | Authors: | Pesce, A, Tilleman, L, Donne, J, Aste, E, Ascenzi, P, Ciaccio, C, Coletta, M, Moens, L, Viappiani, C, Dewilde, S, Bolognesi, M, Nardini, M. | | Deposit date: | 2013-01-18 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure and Haem-Distal Site Plasticity in Methanosarcina Acetivorans Protoglobin.
Plos One, 8, 2013
|
|
3ZMS
 
 | | LSD1-CoREST in complex with INSM1 peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, INSULINOMA-ASSOCIATED PROTEIN 1, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
3ZOM
 
 | | M.acetivorans protoglobin F145W mutant | | Descriptor: | GLYCEROL, PROTOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tilleman, L, Abbruzzetti, S, Ciaccio, C, De Sanctis, G, Nardini, M, Pesce, A, Desmet, F, Moens, L, Van Doorslaer, S, Bruno, S, Bolognesi, M, Ascenzi, P, Coletta, M, Viappiani, C, Dewilde, S. | | Deposit date: | 2013-02-22 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Bases for the Regulation of Co Binding in the Archaeal Protoglobin from Methanosarcina Acetivorans.
Plos One, 10, 2015
|
|
2A87
 
 | | Crystal Structure of M. tuberculosis Thioredoxin reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Akif, M, Suhre, K, Verma, C, Mande, S.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-07-07 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conformational flexibility of Mycobacterium tuberculosis thioredoxin reductase: crystal structure and normal-mode analysis.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
8XX0
 
 | | Crystal structure of anti-IgE antibody HMK-12 Fab complexed with IgE F(ab')2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SPE7 immunoglobulin E F(ab')2 heavy chain, ... | | Authors: | Hirano, T, Koyanagi, A, Kasai, M, Okumura, K. | | Deposit date: | 2024-01-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric inhibition of IgE-Fc epsilon RI interactions by simultaneous targeting of IgE F(ab')2 epitopes.
Commun Biol, 7, 2024
|
|
3ZMT
 
 | | LSD1-CoREST in complex with PRSFLV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
2A0B
 
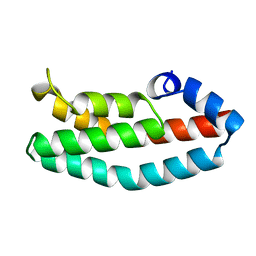 | | HISTIDINE-CONTAINING PHOSPHOTRANSFER DOMAIN OF ARCB FROM ESCHERICHIA COLI | | Descriptor: | HPT DOMAIN, ZINC ION | | Authors: | Kato, M, Mizuno, T, Shimizu, T, Hakoshima, T. | | Deposit date: | 1998-04-02 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Refined structure of the histidine-containing phosphotransfer (HPt) domain of the anaerobic sensor kinase ArcB from Escherichia coli at 1.57 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1HJI
 
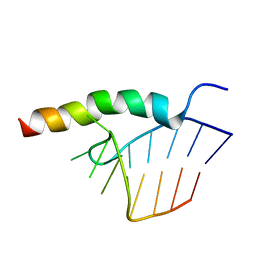 | | BACTERIOPHAGE HK022 NUN-PROTEIN-NUTBOXB-RNA COMPLEX | | Descriptor: | NUN-PROTEIN, RNA (5-R(P*GP*CP*CP*CP*UP*GP*AP*AP*AP*AP*AP*GP*GP*GP*C)-3) | | Authors: | Faber, C, Schaerpf, M, Becker, T, Sticht, H, Roesch, P. | | Deposit date: | 2001-01-15 | | Release date: | 2002-01-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Coliphage Hk022 Nun Protein-Lambda-Phage Boxb RNA Complex. Implications for the Mechanism of Transcription Termination
J.Biol.Chem., 276, 2001
|
|
2ANJ
 
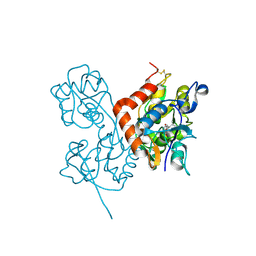 | | Crystal Structure of the Glur2 Ligand Binding Core (S1S2J-Y450W) Mutant in Complex With the Partial Agonist Kainic Acid at 2.1 A Resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2 | | Authors: | Holm, M.M, Naur, P, Vestergaard, B, Geballe, M.T, Gajhede, M, Kastrup, J.S, Traynelis, S.F, Egebjerg, J. | | Deposit date: | 2005-08-11 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Binding Site Tyrosine Shapes Desensitization Kinetics and Agonist Potency at GluR2: a mutagenic, kinetic, and crystallographic study
J.Biol.Chem., 280, 2005
|
|
2ANR
 
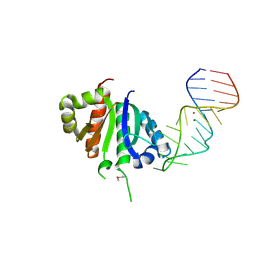 | | Crystal structure (II) of Nova-1 KH1/KH2 domain tandem with 25nt RNA hairpin | | Descriptor: | 5'-R(*CP*(5BU)P*CP*GP*CP*GP*GP*AP*UP*CP*AP*GP*UP*CP*AP*CP*CP*CP*AP*AP*GP*CP*GP*AP*G)-3', MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Malinina, L, Teplova, M, Musunuru, K, Teplov, A, Darnell, J.C, Burley, S.K, Darnell, R.B, Patel, D.J. | | Deposit date: | 2005-08-11 | | Release date: | 2006-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Protein-RNA and protein-protein recognition by dual KH1/2 domains of the neuronal splicing factor Nova-1.
Structure, 19, 2011
|
|
1HPJ
 
 | |
4MLD
 
 | | X-ray structure of ComE D58E REC domain from Streptococcus pneumoniae | | Descriptor: | Response regulator | | Authors: | Boudes, M, Sanchez, D, Durand, D, Graille, M, van Tilbeurgh, H, Quevillon-Cheruel, S. | | Deposit date: | 2013-09-06 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural insights into the dimerization of the response regulator ComE from Streptococcus pneumoniae.
Nucleic Acids Res., 42, 2014
|
|
2AXE
 
 | | IODINATED COMPLEX OF ACETYL XYLAN ESTERASE AT 1.80 ANGSTROMS | | Descriptor: | ACETYL XYLAN ESTERASE, SULFATE ION | | Authors: | Ghosh, D, Erman, M, Sawicki, M.W, Lala, P, Weeks, D.R, Li, N, Pangborn, W, Thiel, D.J, Jornvall, H, Eyzaguirre, J. | | Deposit date: | 1998-09-01 | | Release date: | 1999-05-18 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Determination of a protein structure by iodination: the structure of iodinated acetylxylan esterase.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2ANI
 
 | | Crystal structure of the F127Y mutant of Ribonucleotide Reductase R2 from Chlamydia trachomatis | | Descriptor: | FE (III) ION, LEAD (II) ION, Ribonucleoside-diphosphate reductase beta subunit | | Authors: | Hogbom, M, Stenmark, P, Nordlund, P. | | Deposit date: | 2005-08-11 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the high-valent FeIIIFeIV state in ribonucleotide reductase (RNR) of Chlamydia trachomatis--combined EPR, 57Fe-, 1H-ENDOR and X-ray studies.
Biochim.Biophys.Acta, 1774, 2007
|
|
2B1N
 
 | |
6M04
 
 | | Structure of the human homo-hexameric LRRC8D channel at 4.36 Angstroms | | Descriptor: | Volume-regulated anion channel subunit LRRC8D | | Authors: | Nakamura, R, Kasuya, G, Yokoyama, T, Shirouzu, M, Ishitani, R, Nureki, O. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Cryo-EM structure of the volume-regulated anion channel LRRC8D isoform identifies features important for substrate permeation.
Commun Biol, 3, 2020
|
|
4MYX
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ame complexed with P32 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-5-{[(2-{3-[(1E)-N-hydroxyethanimidoyl]phenyl}propan-2-yl)carbamoyl]amino}benzamide, FORMIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-28 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ame complexed with P32
To be Published
|
|
4MYA
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor A110 | | Descriptor: | 4-{(1R)-1-[1-(4-chlorophenyl)-1H-1,2,3-triazol-4-yl]ethoxy}quinolin-2(1H)-one, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-27 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8997 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor A110
To be Published
|
|
8J47
 
 | | CryoEM Structure of 40-Residue Arctic (E22G) Beta-Amyloid Fibril Derived by Co-Analysis with Solid-State NMR | E22G Abeta40 | | Descriptor: | E22G Amyloid-beta | | Authors: | Tehrani, M.J, Matsuda, I, Yamagata, A, Matsunaga, T, Sato, M, Toyooka, K, Shirouzu, M, Ishii, Y, Kodama, Y, McElheny, D, Kobayashi, N. | | Deposit date: | 2023-04-19 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | E22G A beta 40 fibril structure and kinetics illuminate how A beta 40 rather than A beta 42 triggers familial Alzheimer's.
Nat Commun, 15, 2024
|
|
