7P5T
 
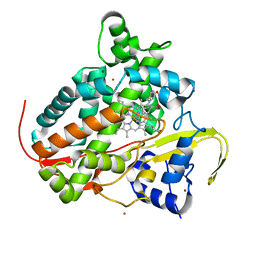 | | Structure of CYP142 from Mycobacterium tuberculosis in complex with inhibitor MEK216 | | Descriptor: | BROMIDE ION, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Snee, M, Kavanagh, M, Tunnicliffe, R, McLean, K, Levy, C, Munro, A. | | Deposit date: | 2021-07-14 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of CYP142 from Mycobacterium tuberculosis in complex with inhibitor MEK216
To Be Published
|
|
4WYZ
 
 | | The crystal structure of the A109G mutant of RNase A in complex with 3'UMP | | Descriptor: | 3'-URIDINEMONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | French, R.L, Gagne, D, Doucet, N, Simonovic, M. | | Deposit date: | 2014-11-18 | | Release date: | 2015-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Perturbation of the Conformational Dynamics of an Active-Site Loop Alters Enzyme Activity.
Structure, 23, 2015
|
|
4WZO
 
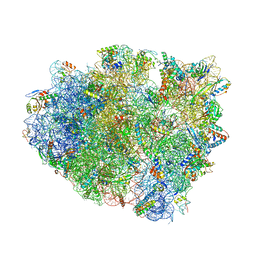 | | Complex of 70S ribosome with tRNA-fMet and mRNA | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2014-11-20 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the translational infidelity mechanism.
Nat Commun, 6, 2015
|
|
4RIL
 
 | | Structure of the amyloid forming segment, GAVVTGVTAVA, from the NAC domain of Parkinson's disease protein alpha-synuclein, residues 68-78, determined by electron diffraction | | Descriptor: | Alpha-synuclein | | Authors: | Rodriguez, J.A, Ivanova, M, Sawaya, M.R, Cascio, D, Reyes, F, Shi, D, Johnson, L, Guenther, E, Sangwan, S, Hattne, J, Nannenga, B, Brewster, A.S, Messerschmidt, M, Boutet, S, Sauter, N.K, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2014-10-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-20 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.43 Å) | | Cite: | Structure of the toxic core of alpha-synuclein from invisible crystals.
Nature, 525, 2015
|
|
7AVB
 
 | |
7AVA
 
 | |
4X5V
 
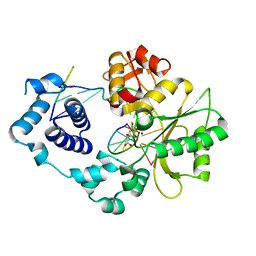 | | Crystal structure of the post-catalytic nick complex of DNA polymerase lambda with a templating A and incorporated 8-oxo-dGMP | | Descriptor: | CITRIC ACID, DNA (5'-D(*CP*AP*GP*TP*AP*CP*(8OG))-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Burak, M.J, Guja, K.E, Garcia-Diaz, M. | | Deposit date: | 2014-12-05 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nucleotide binding interactions modulate dNTP selectivity and facilitate 8-oxo-dGTP incorporation by DNA polymerase lambda.
Nucleic Acids Res., 43, 2015
|
|
5Z3A
 
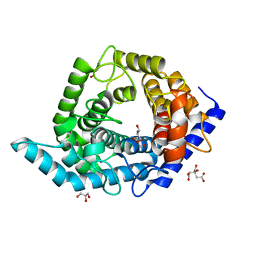 | | Glycosidase Wild Type | | Descriptor: | CITRIC ACID, GLYCEROL, Glycoside hydrolase 15-related protein | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
8SUV
 
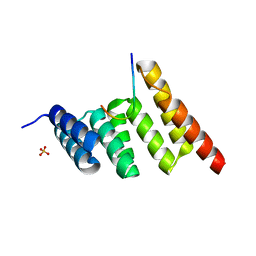 | | CHIP-TPR in complex with the C-terminus of CHIC2 | | Descriptor: | Cysteine-rich hydrophobic domain-containing protein 2, E3 ubiquitin-protein ligase CHIP, SULFATE ION | | Authors: | Cupo, A.R, McDermott, L.E, DeSilva, A.R, Callahan, M, Nix, J.C, Gestwicki, J.E, Page, R.C. | | Deposit date: | 2023-05-13 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Interaction with the membrane-anchored protein CHIC2 constrains the ubiquitin ligase activity of CHIP
Biorxiv, 2023
|
|
4WT1
 
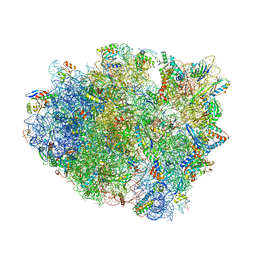 | | Complex of 70S ribosome with tRNA-Phe and mRNA with A-A mismatch in the second position in the A-site | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2014-10-29 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into the translational infidelity mechanism.
Nat Commun, 6, 2015
|
|
4WYN
 
 | |
4X1B
 
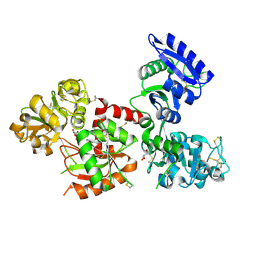 | | Human serum transferrin with ferric ion bound at the C-lobe only | | Descriptor: | FE (III) ION, GLYCEROL, MALONATE ION, ... | | Authors: | Wang, M, Zhang, H, Sun, H. | | Deposit date: | 2014-11-24 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | "Anion clamp" allows flexible protein to impose coordination geometry on metal ions
Chem.Commun.(Camb.), 51, 2015
|
|
4X2M
 
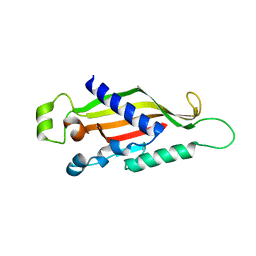 | | Structure of Mtr2 | | Descriptor: | Mtr2 | | Authors: | Aibara, S, Valkov, E, Stewart, M. | | Deposit date: | 2014-11-26 | | Release date: | 2015-07-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of the principal mRNA-export factor Mex67-Mtr2 from Chaetomium thermophilum.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
8HUA
 
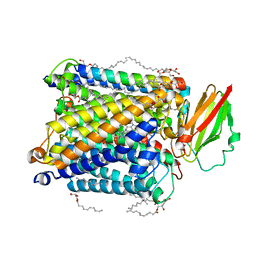 | | Serial synchrotron crystallography structure of ba3-type cytochrome c oxidase from Thermus thermophilus using a goniometer compatible flow-cell | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Ghosh, S, Zoric, D, Bjelcic, M, Johannesson, J, Sandelin, E, Branden, G, Neutze, R. | | Deposit date: | 2022-12-22 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A simple goniometer-compatible flow cell for serial synchrotron X-ray crystallography.
J.Appl.Crystallogr., 56, 2023
|
|
4X69
 
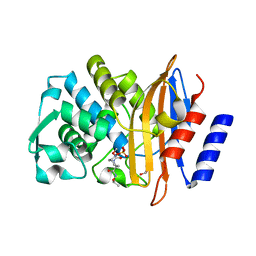 | | Crystal structure of OP0595 complexed with CTX-M-44 | | Descriptor: | (2S,5R)-N-(2-aminoethoxy)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, Beta-lactamase Toho-1 | | Authors: | Yamada, M, Watanabe, T. | | Deposit date: | 2014-12-07 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | OP0595, a new diazabicyclooctane: mode of action as a serine beta-lactamase inhibitor, antibiotic and beta-lactam 'enhancer'
J.Antimicrob.Chemother., 70, 2015
|
|
8SVE
 
 | |
6A3H
 
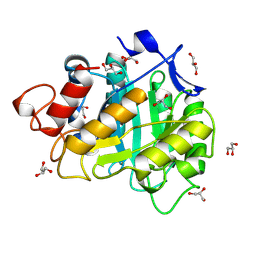 | | 2-DEOXY-2-FLURO-B-D-CELLOTRIOSYL/ENZYME INTERMEDIATE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.6 ANGSTROM RESOLUTION | | Descriptor: | ENDOGLUCANASE, GLYCEROL, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Davies, G.J, Varrot, A, Dauter, M, Brzozowski, A.M, Schulein, M, Mackenzie, L, Withers, S.G. | | Deposit date: | 1998-07-22 | | Release date: | 1999-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase.
Biochemistry, 37, 1998
|
|
6GV8
 
 | |
4X0T
 
 | |
4X0U
 
 | | Structure ALDH7A1 inactivated by 4-diethylaminobenzaldehyde | | Descriptor: | 4-(diethylamino)benzaldehyde, Alpha-aminoadipic semialdehyde dehydrogenase, MAGNESIUM ION | | Authors: | Luo, M, Tanner, J.J. | | Deposit date: | 2014-11-23 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Diethylaminobenzaldehyde Is a Covalent, Irreversible Inactivator of ALDH7A1.
Acs Chem.Biol., 10, 2015
|
|
2R9A
 
 | | Crystal structure of human XLF | | Descriptor: | Non-homologous end-joining factor 1 | | Authors: | Andres, S.N, Junop, M.S. | | Deposit date: | 2007-09-12 | | Release date: | 2008-01-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human XLF: A Twist in Nonhomologous DNA End-Joining
Mol.Cell, 28
|
|
4X2H
 
 | | Sac3N peptide bound to Mex67:Mtr2 | | Descriptor: | Putative mRNA export protein, Putative uncharacterized protein, SER-SER-VAL-PHE-GLY-ALA-PRO-ALA | | Authors: | Aibara, S, Valkov, E, Stewart, M. | | Deposit date: | 2014-11-26 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Characterization of the Chaetomium thermophilum TREX-2 Complex and its Interaction with the mRNA Nuclear Export Factor Mex67:Mtr2.
Structure, 23, 2015
|
|
4X4J
 
 | | Structural and Functional Studies of BexE: Insights into Oxidation During BE-7585A Biosynthesis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative oxygenase, SULFATE ION | | Authors: | Tsai, S.-C, Jackson, D.R, Patel, A, Barajas, J.F, Rohr, J, Yu, X, Liu, H.-W, Sasaki, E, Calveras, J, Metsa-Ketela, M. | | Deposit date: | 2014-12-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Functional Studies of BexE: Insights into Oxidation During BE-7585A Biosynthesis
To Be Published
|
|
7X9U
 
 | | Type-II KH motif of human mitochondrial RbfA | | Descriptor: | Putative ribosome-binding factor A, mitochondrial | | Authors: | Kuwasako, K, Suzuki, S, Furue, M, Takizawa, M, Takahashi, M, Tsuda, K, Nagata, T, Watanabe, S, Tanaka, A, Kobayashi, N, Kigawa, T, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structures of the KH domain of human ribosome binding factor A, mtRbfA, involved in mitochondrial ribosome biogenesis.
Biomol.Nmr Assign., 16, 2022
|
|
4XBA
 
 | | Hnt3 | | Descriptor: | Aprataxin-like protein, GLYCEROL, GUANOSINE, ... | | Authors: | Jacewicz, A, Chauleau, M, Shuman, S. | | Deposit date: | 2014-12-16 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | DNA3'pp5'G de-capping activity of aprataxin: effect of cap nucleoside analogs and structural basis for guanosine recognition.
Nucleic Acids Res., 43, 2015
|
|
