5ULL
 
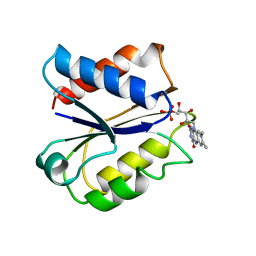 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN: REDUCED | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1997-01-09 | | Release date: | 1997-03-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
8JXF
 
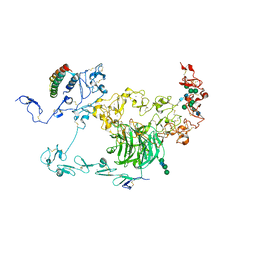 | | rat megalin RAP complex bodyA | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin receptor-associated protein, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7AK0
 
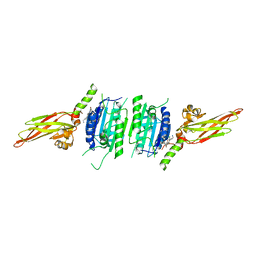 | | Human MALT1(329-729) in complex with a chromane urea containing inhibitor | | Descriptor: | 1-[4-[4-(aminomethyl)pyrazol-1-yl]-3-chloranyl-phenyl]-3-[(3~{R})-6-bromanyl-3,4-dihydro-2~{H}-chromen-3-yl]urea, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.316 Å) | | Cite: | Discovery of Potent, Highly Selective, and In Vivo Efficacious, Allosteric MALT1 Inhibitors by Iterative Scaffold Morphing.
J.Med.Chem., 63, 2020
|
|
4O9R
 
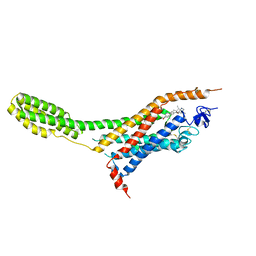 | | Human Smoothened Receptor structure in complex with cyclopamine | | Descriptor: | Cyclopamine, Smoothened homolog/Soluble cytochrome b562 chimeric protein | | Authors: | Wang, C, Weierstall, U, James, D, White, T.A, Wang, D, Liu, W, Spence, J.C.H, Doak, R.B, Nelson, G, Fromme, P, Fromme, R, Grotjohann, I, Kupitz, C, Zatsepin, N.A, Liu, H, Basu, S, Wacker, D, Han, G.W, Katritch, V, Boutet, S, Messerschmidt, M, Willams, G.J, Koglin, J.E, Seibert, M.M, Klinker, M, Gati, C, Shoeman, R.L, Barty, A, Chapman, H.N, Kirian, R.A, Beyerlein, K.R, Stevens, R.C, Li, D, Shah, S.T.A, Howe, N, Caffrey, M, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography.
Nat Commun, 5, 2014
|
|
8JXJ
 
 | | rat megalin RAP complex leg | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LDL receptor related protein 2, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5CZB
 
 | | HCV NS5B IN COMPLEX WITH LIGAND IDX17119-5 | | Descriptor: | 1-[4-(7-amino-5-methylpyrazolo[1,5-a]pyrimidin-2-yl)phenyl]-3-{[(R)-(2,4-dimethylphenyl)(methoxy)phosphoryl]amino}-1H-pyrazole-4-carboxylic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pierra, C, Dousson, C, Augustin, M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-06-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Synthesis of potent and broad genotypically active NS5B HCV non-nucleoside inhibitors binding to the thumb domain allosteric site 2 of the viral polymerase.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
7B33
 
 | | MST3 in complex with MRIA11 | | Descriptor: | 1,2-ETHANEDIOL, 8-[(5-azanyl-1,3-dioxan-2-yl)methyl]-6-[4-[6-[bis(fluoranyl)methyl]pyridin-2-yl]-2-chloranyl-phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
8JXD
 
 | | Cryo-EM structure of rat megalin leg | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QSI
 
 | | human Carbonic Anhydrase II in complex with indoline-1-sulfonamide | | Descriptor: | 2,3-dihydroindole-1-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2022-01-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.304 Å) | | Cite: | Diversely substituted sulfamides for fragment-based drug discovery of carbonic anhydrase inhibitors: synthesis and inhibitory profile.
J Enzyme Inhib Med Chem, 37, 2022
|
|
4INK
 
 | | Crystal structure of SplD protease from Staphylococcus aureus at 1.56 A resolution | | Descriptor: | Serine protease SplD | | Authors: | Zdzalik, M, Kalinska, M, Cichon, P, Wysocka, M, Stec-Niemczyk, J, Stennicke, H.R, Jabaiah, A, Markiewicz, M, Wladyka, B, Daugherty, P.S, Lesner, A, Rolka, K, Dubin, A, Potempa, J, Dubin, G. | | Deposit date: | 2013-01-04 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Biochemical and Structural Characterization of SplD Protease from Staphylococcus aureus.
Plos One, 8, 2013
|
|
8JUT
 
 | | rat megalin RAP complex | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-27 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7AXC
 
 | | Crystal structure of the hPXR-LBD in complex with ferutinine | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Nuclear receptor subfamily 1 group I member 2, ... | | Authors: | Granell, M, Delfosse, V, Bourguet, W. | | Deposit date: | 2020-11-09 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanistic insights into the synergistic activation of the RXR-PXR heterodimer by endocrine disruptor mixtures.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7AXJ
 
 | | Crystal structure of the hPXR-LBD in complex with estradiol and clotrimazole | | Descriptor: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, ESTRADIOL, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Delfosse, V, Granell, M, Blanc, P, Bourguet, W. | | Deposit date: | 2020-11-09 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insights into the synergistic activation of the RXR-PXR heterodimer by endocrine disruptor mixtures.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8JXE
 
 | | rat megalin RAP complex head | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7LHQ
 
 | | Solution structure of SARS-CoV-2 nonstructural protein 7 at pH 7.0 | | Descriptor: | Non-structural protein 7 | | Authors: | Lee, Y, Tonelli, M, Anderson, T.K, Kirchdoerfer, R.N, Henzler-Wildman, K, Lee, W. | | Deposit date: | 2021-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | pH-dependent polymorphism of the structure of SARS-CoV-2 nsp7
Biorxiv, 2021
|
|
7B35
 
 | | MST3 in complex with compound MRIA13 | | Descriptor: | 8-[(5-azanyl-1,3-dioxan-2-yl)methyl]-6-[2-chloranyl-4-(3-methoxy-6-methyl-pyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.40005136 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
4INL
 
 | | Crystal structure of SplD protease from Staphylococcus aureus at 2.1 A resolution | | Descriptor: | Serine protease SplD | | Authors: | Cichon, P, Zdzalik, M, Kalinska, M, Wysocka, M, Stec-Niemczyk, J, Stennicke, H.R, Jabaiah, A, Markiewicz, M, Wladyka, B, Daugherty, P.S, Lesner, A, Rolka, K, Dubin, A, Potempa, J, Dubin, G. | | Deposit date: | 2013-01-04 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and Structural Characterization of SplD Protease from Staphylococcus aureus.
Plos One, 8, 2013
|
|
7B30
 
 | | MST3 in complex with compound G-5555 | | Descriptor: | 8-[(trans-5-amino-1,3-dioxan-2-yl)methyl]-6-[2-chloro-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7(8H)-one, Serine/threonine-protein kinase 24 | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
8JXI
 
 | | rat megalin RAP complex wingB | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXA
 
 | | cryo-EM structure of rat megalin bodyB | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXB
 
 | | Cryo-EM structure of rat megalin wingAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXG
 
 | | rat megalin RAP complex bodyB | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6G7L
 
 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: 8.3 ms state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
8JXC
 
 | | rat megalin wingB | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6G8G
 
 | | Flavonoid-responsive Regulator FrrA in complex with Genistein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GENISTEIN, TetR/AcrR family transcriptional regulator | | Authors: | Werner, N, Hoppen, J, Palm, G, Werten, S, Goettfert, M, Hinrichs, W. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The induction mechanism of the flavonoid-responsive regulator FrrA.
Febs J., 2021
|
|
