7ER7
 
 | | Crystal structure of hyman Biliverdin IX-beta reductase B with Tamibarotene (A80) | | Descriptor: | 4-[(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)carbamoyl]benzoic acid, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7F82
 
 | | Structure of the bacterial cellulose synthase subunit Z in complex with cellooligosaccharides from Enterobacter sp. CJF-002 | | Descriptor: | Glucanase, S,R MESO-TARTARIC ACID, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Fujiwara, T, Fujishima, A, Yao, M. | | Deposit date: | 2021-06-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural snapshot of a glycoside hydrolase family 8 endo-beta-1,4-glucanase capturing the state after cleavage of the scissile bond.
Acta Crystallogr.,Sect.D, 78, 2022
|
|
7ERD
 
 | | Crystal structure of human Biliverdin IX-beta reductase B with Flunixin Meglumin (FMG) | | Descriptor: | 2-[[2-methyl-3-(trifluoromethyl)phenyl]amino]pyridine-3-carboxylic acid, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7Z52
 
 | | Human NEXT dimer - focused reconstruction of the single MTR4 | | Descriptor: | Exosome RNA helicase MTR4, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RNA (5'-R(P*UP*UP*UP*UP*U)-3'), ... | | Authors: | Gerlach, P, Lingaraju, M, Salerno-Kochan, A, Bonneau, F, Basquin, J, Conti, E. | | Deposit date: | 2022-03-07 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and regulation of the nuclear exosome targeting complex guides RNA substrates to the exosome.
Mol.Cell, 82, 2022
|
|
7ER8
 
 | | Crystal structure of human Biliverdin IX-beta reductase B with Sulfasalazine (SAS) | | Descriptor: | 2-HYDROXY-(5-([4-(2-PYRIDINYLAMINO)SULFONYL]PHENYL)AZO)BENZOIC ACID, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7ERB
 
 | | Crystal structure of human Biliverdin IX-beta reductase B with Ataluren (PTC) | | Descriptor: | 3-[5-(2-fluorophenyl)-1,2,4-oxadiazol-3-yl]benzoic acid, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7Z4Z
 
 | | Human NEXT dimer - focused reconstruction of the dimerization module | | Descriptor: | Exosome RNA helicase MTR4, Zinc finger CCHC domain-containing protein 8 | | Authors: | Gerlach, P, Lingaraju, M, Salerno-Kochan, A, Bonneau, F, Basquin, J, Conti, E. | | Deposit date: | 2022-03-06 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and regulation of the nuclear exosome targeting complex guides RNA substrates to the exosome.
Mol.Cell, 82, 2022
|
|
3BP1
 
 | | Crystal structure of putative 7-cyano-7-deazaguanine reductase QueF from Vibrio cholerae O1 biovar eltor | | Descriptor: | GUANINE, MAGNESIUM ION, NADPH-dependent 7-cyano-7-deazaguanine reductase, ... | | Authors: | Kim, Y, Zhou, M, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-18 | | Release date: | 2008-01-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | High-resolution structure of the nitrile reductase QueF combined with molecular simulations provide insight into enzyme mechanism.
J.Mol.Biol., 404, 2010
|
|
7F81
 
 | | Structure of the bacterial cellulose synthase subunit Z from Enterobacter sp. CJF-002 | | Descriptor: | GLYCEROL, Glucanase, S,R MESO-TARTARIC ACID | | Authors: | Fujiwara, T, Fujishima, A, Yao, M. | | Deposit date: | 2021-06-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural snapshot of a glycoside hydrolase family 8 endo-beta-1,4-glucanase capturing the state after cleavage of the scissile bond.
Acta Crystallogr.,Sect.D, 78, 2022
|
|
7Z4Y
 
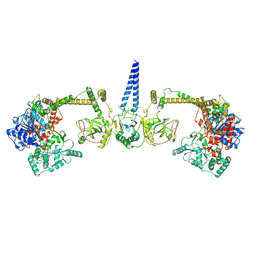 | | Human NEXT dimer - overall reconstruction of the core complex | | Descriptor: | Exosome RNA helicase MTR4, Zinc finger CCHC domain-containing protein 8 | | Authors: | Gerlach, P, Lingaraju, M, Salerno-Kochan, A, Bonneau, F, Basquin, J, Conti, E. | | Deposit date: | 2022-03-06 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and regulation of the nuclear exosome targeting complex guides RNA substrates to the exosome.
Mol.Cell, 82, 2022
|
|
7ESI
 
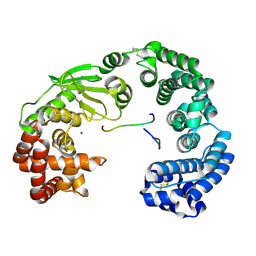 | | Crystal structure of the collagenase unit of a Vibrio collagenase from Vibrio harveyi VHJR7 at 1. 8 angstrom resolution. | | Descriptor: | CALCIUM ION, Collagenase unit (CU), Peptide P1, ... | | Authors: | Cao, H.Y, Wang, Y, Peng, M, Zhang, Y.Z. | | Deposit date: | 2021-05-11 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Vibrio collagenase VhaC provides insight into the mechanism of bacterial collagenolysis.
Nat Commun, 13, 2022
|
|
3BRE
 
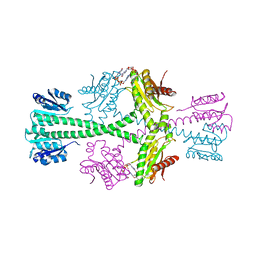 | | Crystal Structure of P.aeruginosa PA3702 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, Probable two-component response regulator | | Authors: | De, N, Pirruccello, M, Krasteva, P.V, Bae, N, Raghavan, R.V, Sondermann, H. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phosphorylation-independent regulation of the diguanylate cyclase WspR.
Plos Biol., 6, 2008
|
|
3CYZ
 
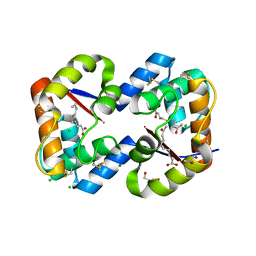 | | Dimeric crystal structure of a pheromone binding protein from Apis mellifera in complex with 9-keto-2(E)-decenoic acid at pH 7.0 | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-04-27 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
7Z4M
 
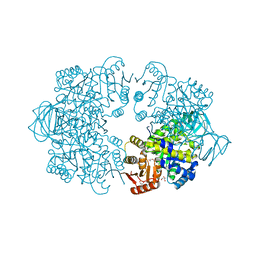 | | Plasmodium falciparum pyruvate kinase complexed with Mg2+ and K+ | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Dillenberger, M, Rahlfs, S, Becker, K, Fritz-Wolf, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Prominent role of cysteine residues C49 and C343 in regulating Plasmodium falciparum pyruvate kinase activity.
Structure, 30, 2022
|
|
7Z4R
 
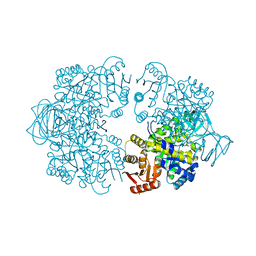 | | Plasmodium falciparum pyruvate kinase mutant - C343A | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, Pyruvate kinase | | Authors: | Dillenberger, M, Rahlfs, S, Becker, K, Fritz-Wolf, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prominent role of cysteine residues C49 and C343 in regulating Plasmodium falciparum pyruvate kinase activity.
Structure, 30, 2022
|
|
7Z4N
 
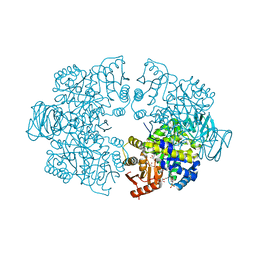 | | Plasmodium falciparum pyruvate kinase complexed with pyruvate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Dillenberger, M, Rahlfs, S, Becker, K, Fritz-Wolf, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Prominent role of cysteine residues C49 and C343 in regulating Plasmodium falciparum pyruvate kinase activity.
Structure, 30, 2022
|
|
7Z4Q
 
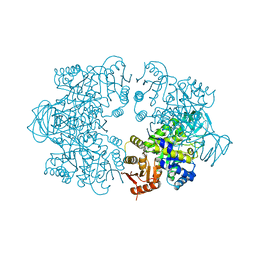 | | Plasmodium falciparum pyruvate kinase mutant - C49A | | Descriptor: | MAGNESIUM ION, Pyruvate kinase | | Authors: | Dillenberger, M, Rahlfs, S, Becker, K, Fritz-Wolf, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Prominent role of cysteine residues C49 and C343 in regulating Plasmodium falciparum pyruvate kinase activity.
Structure, 30, 2022
|
|
3D79
 
 | | Crystal structure of hypothetical protein PH0734.1 from hyperthermophilic archaea Pyrococcus horikoshii OT3 | | Descriptor: | Putative uncharacterized protein PH0734 | | Authors: | Nishimura, Y, Miyazono, K, Sawano, Y, Makino, T, Nagata, K, Tanokura, M. | | Deposit date: | 2008-05-20 | | Release date: | 2008-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of hypothetical protein PH0734.1 from hyperthermophilic archaea Pyrococcus horikoshii OT3.
Proteins, 73, 2008
|
|
3D7L
 
 | |
7Z1L
 
 | | Structure of yeast RNA Polymerase III Pre-Termination Complex (PTC) | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7YZX
 
 | | ScpA from Streptococcus pyogenes, D783A mutant. | | Descriptor: | C5a peptidase, CALCIUM ION, MALONIC ACID, ... | | Authors: | Kagawa, T.F, Cooney, J.C, Jain, M. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray diffraction data for the C5a-peptidase mutant with modified activity and specificity.
Data Brief, 46, 2023
|
|
7YL9
 
 | | Cryo-EM structure of complete transmembrane channel E289A mutant Vibrio cholerae Cytolysin | | Descriptor: | Hemolysin | | Authors: | Mondal, A.K, Sengupta, N, Singh, M, Biswas, R, Lata, K, Lahiri, I, Dutta, S, Chattopadhyay, K. | | Deposit date: | 2022-07-25 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM structure of complete transmembrane channel E289A mutant Vibrio cholerae Cytolysin
J.Biol.Chem.
|
|
3CFR
 
 | | Structure of the replicating complex of a POL Alpha family DNA Polymerase, ternary complex 2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*DGP*DCP*DGP*DGP*DAP*DCP*DTP*DGP*DCP*DTP*DTP*DAP*(DOC))-3'), ... | | Authors: | Wang, J, Klimenko, D, Wang, M, Steitz, T.A, Konigsberg, W.H. | | Deposit date: | 2008-03-04 | | Release date: | 2009-03-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into base selectivity from the structures
of an RB69 DNA Polymerase triple mutant
To be Published
|
|
3CFY
 
 | | Crystal structure of signal receiver domain of putative Luxo repressor protein from Vibrio parahaemolyticus | | Descriptor: | Putative LuxO repressor protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Fong, R, Freeman, J, Iizuka, M, Groshong, C, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Putative Luxo Repressor Protein from Vibrio Parahaemolyticus.
To be Published
|
|
7FIN
 
 | | Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GIPR-Gs complex | | Descriptor: | CHOLESTEROL, GAMMA-L-GLUTAMIC ACID, Gastric inhibitory polypeptide receptor,human glucose-dependent insulinotropic polypeptide receptor, ... | | Authors: | Zhao, F.h, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-07-31 | | Release date: | 2022-02-23 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
