7OUC
 
 | |
3BSU
 
 | | Hybrid-binding domain of human RNase H1 in complex with 12-mer RNA/DNA | | Descriptor: | DNA (5'-D(*DGP*DAP*DAP*DTP*DCP*DAP*DGP*DGP*(5IU)P*DGP*DTP*DC)-3'), MAGNESIUM ION, RNA (5'-R(*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*C)-3'), ... | | Authors: | Nowotny, M, Cerritelli, S.M, Ghirlando, R, Gaidamakov, S.A, Crouch, R.J, Yang, W. | | Deposit date: | 2007-12-26 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Specific recognition of RNA/DNA hybrid and enhancement of human RNase H1 activity by HBD.
Embo J., 27, 2008
|
|
3BTT
 
 | | THE CRYSTAL STRUCTURES OF THE COMPLEXES BETWEEN BOVINE BETA-TRYPSIN AND TEN P1 VARIANTS OF BPTI | | Descriptor: | CALCIUM ION, PROTEIN (PANCREATIC TRYPSIN INHIBITOR), PROTEIN (TRYPSIN), ... | | Authors: | Helland, R, Otlewski, J, Sundheim, O, Dadlez, M, Smalas, A.O. | | Deposit date: | 1999-03-10 | | Release date: | 2000-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of the complexes between bovine beta-trypsin and ten P1 variants of BPTI.
J.Mol.Biol., 287, 1999
|
|
7OUD
 
 | |
2A35
 
 | | 1.5 A Crystal Structure of a Protein of Unknown Function PA4017 from Pseudomonas aeruginosa PAO1, Possible Epimerase | | Descriptor: | hypothetical protein PA4017 | | Authors: | Zhang, R, Xu, L, Cuff, M, Savchenko, A, Cymborowski, M, Minor, W, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-06-23 | | Release date: | 2005-08-09 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein PA4017 from
Pseudomonas aeruginosa PAO1
To be Published
|
|
2A25
 
 | | Crystal structure of Siah1 SBD bound to the peptide EKPAAVVAPITTG from SIP | | Descriptor: | Calcyclin-binding protein peptide, Ubiquitin ligase SIAH1, ZINC ION | | Authors: | Santelli, E, Leone, M, Li, C, Fukushima, T, Preece, N.E, Olson, A.J, Ely, K.R, Reed, J.C, Pellecchia, M, Liddington, R.C, Matsuzawa, S. | | Deposit date: | 2005-06-21 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of Siah1-Siah-interacting Protein Interactions and Insights into the Assembly of an E3 Ligase Multiprotein Complex
J.Biol.Chem., 280, 2005
|
|
7P06
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in outward-facing conformation with ADP-orthovanadate/ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADP ORTHOVANADATE, MAGNESIUM ION, ... | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P05
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation with ADP/ATP and rhodamine 6G | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Pleiotropic ABC efflux transporter of multiple drugs, ... | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P03
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation without nucleotides | | Descriptor: | Pleiotropic ABC efflux transporter of multiple drugs | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P04
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation with ADP/ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Pleiotropic ABC efflux transporter of multiple drugs | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
3C9E
 
 | | Crystal structure of the cathepsin K : chondroitin sulfate complex. | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, CALCIUM ION, Cathepsin K, ... | | Authors: | Kienetz, M, Cherney, M.M, James, M.N.G, Bromme, D. | | Deposit date: | 2008-02-15 | | Release date: | 2008-08-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal and molecular structures of a cathepsin K:chondroitin sulfate complex.
J.Mol.Biol., 383, 2008
|
|
7PBK
 
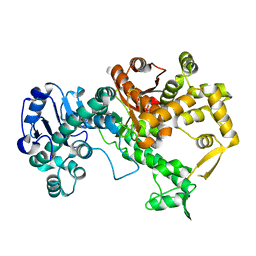 | | Vibriophage phiVC8 family A DNA polymerase (DpoZ), two conformations: thumb-exo open and thumb-exo closed | | Descriptor: | DNA polymerase I | | Authors: | Czernecki, D, Hu, H, Romoli, F, Delarue, M. | | Deposit date: | 2021-08-02 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural dynamics and determinants of 2-aminoadenine specificity in DNA polymerase DpoZ of vibriophage phi VC8.
Nucleic Acids Res., 49, 2021
|
|
3C4O
 
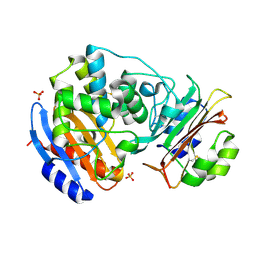 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) E73M/S130K/S146M complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein, SULFATE ION | | Authors: | Reynolds, K.A, Hanes, M.S, Thomson, J.M, Antczak, A.J, Berger, J.M, Bonomo, R.A, Kirsch, J.F, Handel, T.M. | | Deposit date: | 2008-01-30 | | Release date: | 2008-05-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational redesign of the SHV-1 beta-lactamase/beta-lactamase inhibitor protein interface.
J.Mol.Biol., 382, 2008
|
|
3C5K
 
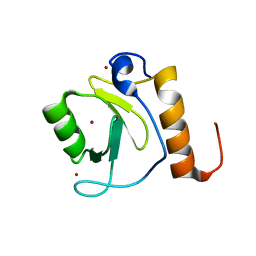 | | Crystal structure of human HDAC6 zinc finger domain | | Descriptor: | Histone deacetylase 6, ZINC ION | | Authors: | Dong, A, Ravichandran, M, Schuetz, A, Loppnau, P, Li, Y, MacKenzie, F, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Dhe-Paganon, S, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Human HDAC6 zinc finger domain.
To be Published
|
|
2AEI
 
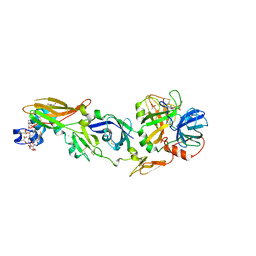 | | Crystal structure of a ternary complex of factor VIIa/tissue factor and 2-[[6-[3-(aminoiminomethyl)phenoxy]-3,5-difluro-4-[(1-methyl-3-phenylpropyl)amino]-2-pyridinyl]oxy]-benzoic acid | | Descriptor: | 2-({6-{3-[AMINO(IMINO)METHYL]PHENOXY}-3,5-DIFLUORO-4-[(1-METHYL-3-PHENYLPROPYL)AMINO]-2-PYRIDINYL}OXY)BENZOIC ACID, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Adler, M, Whitlow, M. | | Deposit date: | 2005-07-22 | | Release date: | 2006-08-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The discovery of fluoropyridine-based inhibitors of the Factor VIIa/TF complex.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
3BP1
 
 | | Crystal structure of putative 7-cyano-7-deazaguanine reductase QueF from Vibrio cholerae O1 biovar eltor | | Descriptor: | GUANINE, MAGNESIUM ION, NADPH-dependent 7-cyano-7-deazaguanine reductase, ... | | Authors: | Kim, Y, Zhou, M, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-18 | | Release date: | 2008-01-08 | | Last modified: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | High-resolution structure of the nitrile reductase QueF combined with molecular simulations provide insight into enzyme mechanism.
J.Mol.Biol., 404, 2010
|
|
3BRE
 
 | | Crystal Structure of P.aeruginosa PA3702 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, Probable two-component response regulator | | Authors: | De, N, Pirruccello, M, Krasteva, P.V, Bae, N, Raghavan, R.V, Sondermann, H. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phosphorylation-independent regulation of the diguanylate cyclase WspR.
Plos Biol., 6, 2008
|
|
7CPZ
 
 | | Crystal structure of Streptoavidin-C1 from Streptomyces cinamonensis | | Descriptor: | BIOTIN, Mature Streptoavidin-C1 | | Authors: | Jeon, B.J, Kim, S, Lee, J.-H, Kim, M.S, Hwang, K.Y. | | Deposit date: | 2020-08-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the structure of mature streptavidin C1 from Streptomyces cinnamonensis reveal the self-binding of the extension C-terminal peptide to biotin-binding sites.
Iucrj, 8, 2021
|
|
7CQ0
 
 | | Crystal structure of Streptoavidin-C1 from Streptomyces cinamonensis | | Descriptor: | Mature Streptoavidin-C1 | | Authors: | Jeon, B.J, Kim, S, Lee, J.-H, Kim, M.S, Hwang, K.Y. | | Deposit date: | 2020-08-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Insights into the structure of mature streptavidin C1 from Streptomyces cinnamonensis reveal the self-binding of the extension C-terminal peptide to biotin-binding sites.
Iucrj, 8, 2021
|
|
2A18
 
 | | carboxysome shell protein ccmK4, crystal form 2 | | Descriptor: | AMMONIUM ION, Carbon dioxide concentrating mechanism protein ccmK homolog 4 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Tanaka, S, Nguyen, C.V, Phillips, M, Beeby, M, Yeates, T.O. | | Deposit date: | 2005-06-18 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Protein structures forming the shell of primitive bacterial organelles
Science, 309, 2005
|
|
7P0L
 
 | |
7Y8P
 
 | | Crystal structure of 4'-selenoRNA duplex | | Descriptor: | COBALT HEXAMMINE(III), RNA (5'-R(*GP*GP*AP*(IKS)P*(ILK)P*(IKS)P*GP*AP*GP*UP*CP*C)-3') | | Authors: | Kondo, J, Minakawa, N, Ohta, M, Takahashi, H, Tarashima, N. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and properties of fully-modified 4'-selenoRNA, an endonuclease-resistant RNA analog.
Bioorg.Med.Chem., 76, 2022
|
|
3BV9
 
 | | Structure of Thrombin Bound to the Inhibitor FM19 | | Descriptor: | FM19 inhibitor, GLYCEROL, IODIDE ION, ... | | Authors: | Nieman, M.T, Burke, F, Warnock, M, Zhou, Y, Sweigert, J, Chen, A, Ricketts, D, Lucchesi, B.R, Chen, Z, Di Cera, E, Hilfinger, J, Mosberg, H.I, Schmaier, A.H. | | Deposit date: | 2008-01-05 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thrombostatin FM compounds: direct thrombin inhibitors - mechanism of action in vitro and in vivo.
J.Thromb.Haemost., 6, 2008
|
|
3CCT
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-[2-(4-fluorophenyl)-4-[(2-hydroxyphenyl)carbamoyl]-5-(1-methylethyl)-3-phenyl-1H-pyrrol-1-yl]-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
3CCZ
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-[2-(4-fluorophenyl)-4-{[(1S)-2-hydroxy-1-phenylethyl]carbamoyl}-5-(1-methylethyl)-1H-imidazol-1-yl]-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
