1RWQ
 
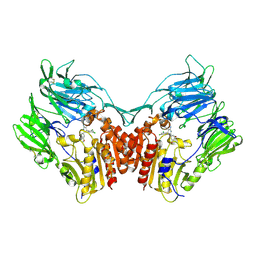 | | Human Dipeptidyl peptidase IV in complex with 5-aminomethyl-6-(2,4-dichloro-phenyl)-2-(3,5-dimethoxy-phenyl)-pyrimidin-4-ylamine | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-(AMINOMETHYL)-6-(2,4-DICHLOROPHENYL)-2-(3,5-DIMETHOXYPHENYL)PYRIMIDIN-4-AMINE, Dipeptidyl peptidase IV | | 著者 | Hennig, M, Thoma, R, Stihle, M. | | 登録日 | 2003-12-17 | | 公開日 | 2004-12-17 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Aminomethylpyrimidines as novel DPP-IV inhibitors: a 10(5)-fold activity increase by optimization of aromatic substituents
Bioorg.Med.Chem.Lett., 14, 2004
|
|
6GHC
 
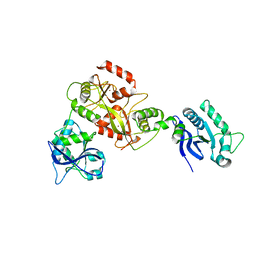 | | Modification dependent EcoKMcrA restriction endonuclease | | 分子名称: | 5-methylcytosine-specific restriction enzyme A, ZINC ION | | 著者 | Czapinska, H, Kowalska, M, Zagorskaite, E, Manakova, E, Xu, S, Siksnys, V, Sasnauskas, G, Bochtler, M. | | 登録日 | 2018-05-07 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Activity and structure of EcoKMcrA.
Nucleic Acids Res., 46, 2018
|
|
1KZ4
 
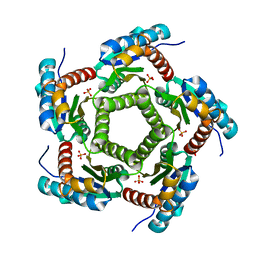 | | Mutant enzyme W63Y Lumazine Synthase from S.pombe | | 分子名称: | 6,7-Dimethyl-8-ribityllumazine Synthase, PHOSPHATE ION | | 著者 | Gerhardt, S, Haase, I, Steinbacher, S, Kaiser, J.T, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | 登録日 | 2002-02-06 | | 公開日 | 2002-07-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase.
J.Mol.Biol., 318, 2002
|
|
8GSX
 
 | |
8G54
 
 | |
8GB2
 
 | | Crystal structure of Apo-SAMHD1 | | 分子名称: | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION | | 著者 | Egleston, M, Dong, L, Howlader, A.H, Bhat, S, Orris, B, Bianchet, M.A, Greenberg, M.M, Stivers, J.T. | | 登録日 | 2023-02-24 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | Deoxyguanosine-Linked Bifunctional Inhibitor of SAMHD1 dNTPase Activity and Nucleic Acid Binding.
Acs Chem.Biol., 18, 2023
|
|
6T2N
 
 | | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown | | 分子名称: | CALCIUM ION, Glycoside hydrolase family 16 protein | | 著者 | Crouch, L.I, Liberato, M.V, Ubranowicz, P.A, Basle, A, Lamb, C.A, Cooke, K, Doona, M, Needham, S, Brady, R.R, Berrington, J.E, Madubic, K, Chater, P, Zhang, F, Linhardt, R.J, Spence, D.I.R, Bolam, D.N. | | 登録日 | 2019-10-09 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown.
Nat Commun, 11, 2020
|
|
3NFR
 
 | | Casimiroin analog inhibitor of quinone reductase 2 | | 分子名称: | 6,8-dimethoxy-1,4-dimethylquinolin-2(1H)-one, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | 著者 | Sturdy, M, Mesecar, A.D, Jermihov, K, Cushman, M, Maiti, A. | | 登録日 | 2010-06-10 | | 公開日 | 2012-01-11 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | X-ray Crystallographic Structure Activity Relationship (SAR) of Casimiroin and its Analogs Bound to Human Quinone Reductase 2
To be Published
|
|
6T3K
 
 | |
5ZK9
 
 | | Stapled-peptides tailored against initiation of translation | | 分子名称: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, ACE-ARG-ILE-ILE-TYR-SER-ARG-MK8-GLN-LEU-LEU-MK8-LEU-LYS-NH2, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Ciesielski, F, Uhring, M. | | 登録日 | 2018-03-23 | | 公開日 | 2019-02-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
8B0I
 
 | | CryoEM structure of bacterial RapZ.GlmZ complex central to the control of cell envelope biogenesis | | 分子名称: | GlmZ small regulatory RNA, RNase adapter protein RapZ | | 著者 | Islam, M.S, Hardwick, H.W, Chirgadze, D.Y, Luisi, B.F. | | 登録日 | 2022-09-07 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.28 Å) | | 主引用文献 | Structure of a bacterial ribonucleoprotein complex central to the control of cell envelope biogenesis.
Embo J., 42, 2023
|
|
8GOB
 
 | | Crystal Structure of Glycerol Dehydrogenase in the presence of NAD+ | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycerol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Park, T, Hoang, H.N, Kang, J.Y, Park, J, Mun, S.A, Jin, M, Yang, J, Jung, C.-H, Eom, S.H. | | 登録日 | 2022-08-24 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural and functional insights into the flexible beta-hairpin of glycerol dehydrogenase.
Febs J., 290, 2023
|
|
6JYP
 
 | | Crystal structure of uPA_H99Y in complex with 3-azanyl-5-(azepan-1-yl)-N-[bis(azanyl)methylidene]-6-chloranyl-pyrazine-2-carboxamide | | 分子名称: | 3-azanyl-5-(azepan-1-yl)-N-[bis(azanyl)methylidene]-6-chloranyl-pyrazine-2-carboxamide, SULFATE ION, Urokinase-type plasminogen activator | | 著者 | Buckley, B, Jiang, L.G, Huang, M.D, Kelso, M, Ranson, M. | | 登録日 | 2019-04-27 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | uPA-HMA
To Be Published
|
|
6GGL
 
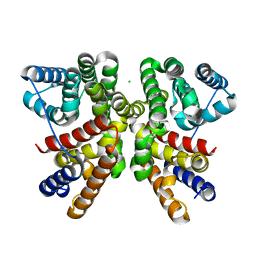 | | Crystal structure of CotB2 variant F107A | | 分子名称: | CHLORIDE ION, Cyclooctat-9-en-7-ol synthase, MAGNESIUM ION | | 著者 | Driller, R, Janke, S, Fuchs, M, Warner, E, Mhashal, A.R, Major, D.T, Christmann, M, Brueck, T, Loll, B. | | 登録日 | 2018-05-03 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Towards a comprehensive understanding of the structural dynamics of a bacterial diterpene synthase during catalysis.
Nat Commun, 9, 2018
|
|
6Y38
 
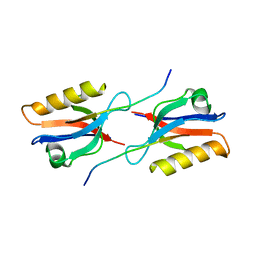 | | Crystal structure of Whirlin PDZ3 in complex with Myosin 15a C-terminal PDZ binding motif peptide | | 分子名称: | Chains: C,D, Whirlin | | 著者 | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | 登録日 | 2020-02-17 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.697 Å) | | 主引用文献 | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
1KY9
 
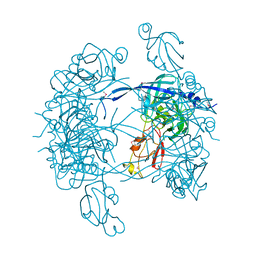 | | Crystal Structure of DegP (HtrA) | | 分子名称: | PROTEASE DO | | 著者 | Krojer, T, Garrido-Franco, M, Huber, R, Ehrmann, M, Clausen, T. | | 登録日 | 2002-02-04 | | 公開日 | 2002-04-03 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine.
Nature, 416, 2002
|
|
6K0H
 
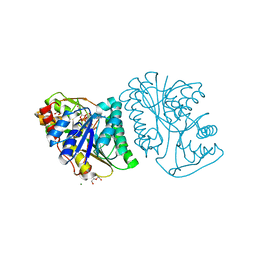 | | Crystal Structure of UDP-glucose 4-epimerase from Bifidobacterium longum in complex with NAD+ and UDP-GlcNAc | | 分子名称: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Nam, Y.-W, Nishimoto, M, Arakawa, T, Kitaoka, M, Fushinobu, S. | | 登録日 | 2019-05-06 | | 公開日 | 2019-08-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for broad substrate specificity of UDP-glucose 4-epimerase in the human milk oligosaccharide catabolic pathway of Bifidobacterium longum.
Sci Rep, 9, 2019
|
|
8GOA
 
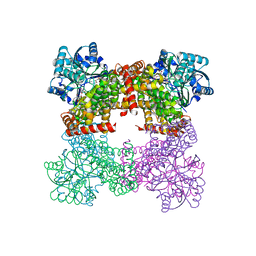 | | Crystal Structure of Glycerol Dehydrogenase in the absence of NAD+ | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycerol dehydrogenase, ZINC ION | | 著者 | Park, T, Hoang, H.N, Kang, J.Y, Park, J, Mun, S.A, Jin, M, Yang, J, Jung, C.-H, Eom, S.H. | | 登録日 | 2022-08-24 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural and functional insights into the flexible beta-hairpin of glycerol dehydrogenase.
Febs J., 290, 2023
|
|
4L7H
 
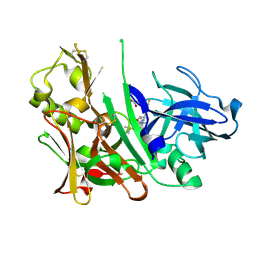 | | Diethylaminosulfur Trifluoride-Mediated Intramolecular Cyclization of 2-hydroxy-benzylureas to Fused Bicyclic Aminooxazoline Compounds and Evaluation of Their Biochemical Activity Against Beta-Secretase-1 (BACE-1) | | 分子名称: | 2-[(3aR,7aR)-2-amino-7a-(2,4-difluorophenyl)-3a,6,7,7a-tetrahydro[1,3]oxazolo[5,4-c]pyridin-5(4H)-yl]pyridine-3-carbonitrile, ACETATE ION, Beta-secretase 1, ... | | 著者 | Huestis, M.P, Liu, W, Volgraf, M, Purkey, H.E, Wang, W, Yu, C, Wu, P, Smith, D, Vigers, G, Dutcher, D, Geck Do, M.K, Hunt, K.W, Siu, M. | | 登録日 | 2013-06-13 | | 公開日 | 2013-09-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Diethylaminosulfur Trifluoride-Mediated Intramolecular Cyclization of 2-hydroxycycloalkylureas to Fused Bicyclic Aminooxazoline Compounds and Evaluation of Their Biochemical Activity Against β-Secretase-1 (BACE-1)
Tetrahedron Lett., 2013
|
|
6YLM
 
 | | mCherry | | 分子名称: | CHLORIDE ION, mCherry | | 著者 | Myskova, J, Rybakova, M, Brynda, J, Lazar, J. | | 登録日 | 2020-04-07 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Directionality of light absorption and emission in representative fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8GH7
 
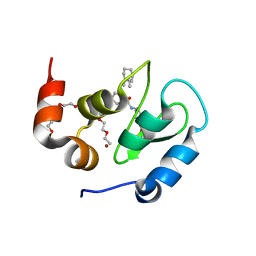 | | 142D6 bound to BIR3-XIAP | | 分子名称: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, BIR3 inhibitor MAA-CHG-PRO-ZHW, E3 ubiquitin-protein ligase XIAP, ... | | 著者 | Garza-Granados, A, McGuire, J, Baggio, C, Pellecchia, M, Pegan, S.D. | | 登録日 | 2023-03-09 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-07-12 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Characterization of a Potent and Orally Bioavailable Lys-Covalent Inhibitor of Apoptosis Protein (IAP) Antagonist.
J.Med.Chem., 66, 2023
|
|
8B0J
 
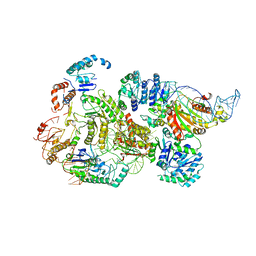 | | CryoEM structure of bacterial RNaseE.RapZ.GlmZ complex central to the control of cell envelope biogenesis | | 分子名称: | GlmZ small RNA, RNase adapter protein RapZ, Ribonuclease E | | 著者 | Islam, M.S, Hardwick, H.W, Chirgadze, D.Y, Luisi, B.F. | | 登録日 | 2022-09-07 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.99 Å) | | 主引用文献 | Structure of a bacterial ribonucleoprotein complex central to the control of cell envelope biogenesis.
Embo J., 42, 2023
|
|
1KYV
 
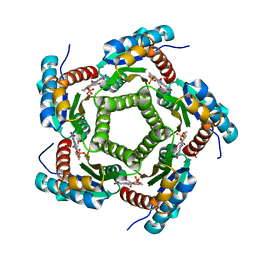 | | Lumazine Synthase from S.pombe bound to riboflavin | | 分子名称: | 6,7-Dimethyl-8-ribityllumazine Synthase, PHOSPHATE ION, RIBOFLAVIN | | 著者 | Gerhardt, S, Haase, I, Steinbacher, S, Kaiser, J.T, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | 登録日 | 2002-02-06 | | 公開日 | 2002-07-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase.
J.Mol.Biol., 318, 2002
|
|
6GCW
 
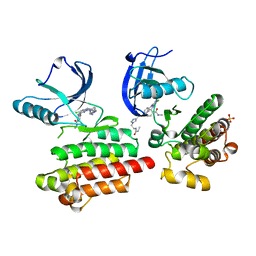 | | Focal Adhesion Kinase catalytic domain in complex with irreversible inhibitor | | 分子名称: | 2-[[5-chloranyl-2-[[4-[[[1-[2-(propanoylamino)ethyl]-1,2,3-triazol-4-yl]methylamino]methyl]phenyl]amino]pyrimidin-4-yl]amino]-~{N}-methyl-benzamide, Focal adhesion kinase 1, SULFATE ION | | 著者 | Yen-Pon, E, Li, B, Acebron-Garcia-de-Eulate, M, Tomkiewicz-Raulet, C, Dawson, J, Lietha, D, Frame, M.C, Coumoul, X, Garbay, C, Etheve-Quelquejeu, M, Chen, H. | | 登録日 | 2018-04-19 | | 公開日 | 2019-05-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Based Design, Synthesis, and Characterization of the First Irreversible Inhibitor of Focal Adhesion Kinase.
Acs Chem.Biol., 13, 2018
|
|
8GKH
 
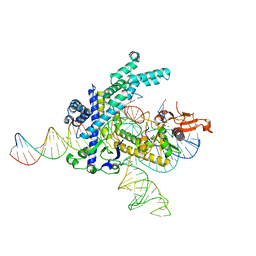 | | Structure of the Spizellomyces punctatus Fanzor (SpuFz) in complex with omega RNA and target DNA | | 分子名称: | DNA (35-MER), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2023-03-19 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Fanzor is a eukaryotic programmable RNA-guided endonuclease.
Nature, 620, 2023
|
|
