8U01
 
 | | Crystal Structure of the Glycoside Hydrolase Family 2 TIM Barrel-domain Containing Protein from Phocaeicola plebeius | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | 著者 | Kim, Y, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2023-08-28 | | 公開日 | 2023-09-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal Structure of the Glycoside Hydrolase Family 2 TIM Barrel-domain
Containing Protein from Phocaeicola plebeius
To Be Published
|
|
8U12
 
 | |
6RY0
 
 | |
6OZ2
 
 | |
9FQH
 
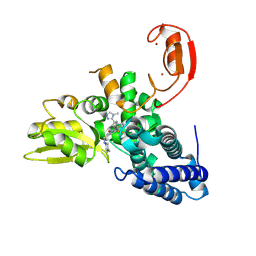 | | E3 ligase Cbl-b in complex with a triazolone core inhibitor (compound 1) | | 分子名称: | 8-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-3-[[(3~{S})-3-methylpiperidin-1-yl]methyl]-5-(trifluoromethyl)-1$l^{4},7,8-triazabicyclo[4.3.0]nona-1(6),2,4-trien-9-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | 著者 | Schimpl, M. | | 登録日 | 2024-06-17 | | 公開日 | 2024-07-31 | | 実験手法 | X-RAY DIFFRACTION (1.786 Å) | | 主引用文献 | Accelerated Discovery of a Carbamate Scaffold Cbl-b Inhibitor using Generative Models and Structure-Based Drug Design
To be published
|
|
9FQJ
 
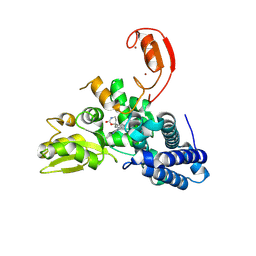 | | E3 ligase Cbl-b in complex with a carbamate scaffold inhibitor (compound 12) | | 分子名称: | 2-cyclopropyl-6-methyl-~{N}-[3-[(6~{S})-6-methyl-2-oxidanylidene-1,3-oxazinan-6-yl]phenyl]pyrimidine-4-carboxamide, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | 著者 | Schimpl, M. | | 登録日 | 2024-06-17 | | 公開日 | 2024-07-31 | | 実験手法 | X-RAY DIFFRACTION (1.563 Å) | | 主引用文献 | Accelerated Discovery of a Carbamate Scaffold Cbl-b Inhibitor using Generative Models and Structure-Based Drug Design
To be published
|
|
8UFM
 
 | | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2 | | 分子名称: | ACETATE ION, FORMIC ACID, Papain-like protease nsp3, ... | | 著者 | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2023-10-04 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2
To Be Published
|
|
8UFL
 
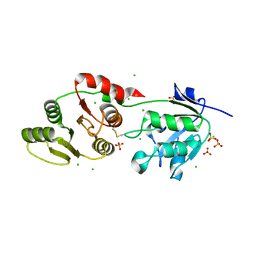 | | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus | | 分子名称: | CHLORIDE ION, Papain-like protease nsp3, SULFATE ION | | 著者 | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2023-10-04 | | 公開日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus
To Be Published
|
|
4O1G
 
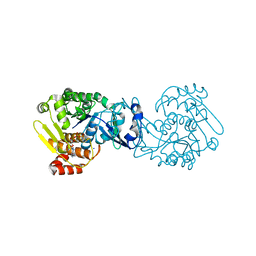 | | MTB adenosine kinase in complex with gamma-Thio-ATP | | 分子名称: | Adenosine kinase, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, SODIUM ION | | 著者 | Dostal, J, Brynda, J, Hocek, M, Pichova, I. | | 登録日 | 2013-12-15 | | 公開日 | 2014-11-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural Basis for Inhibition of Mycobacterial and Human Adenosine Kinase by 7-Substituted 7-(Het)aryl-7-deazaadenine Ribonucleosides
J.Med.Chem., 57, 2014
|
|
8UM3
 
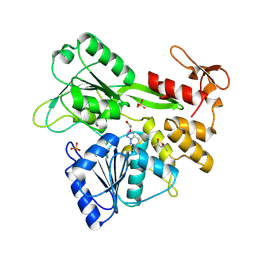 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z203039992 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 6-chlorotetrazolo[1,5-b]pyridazine, ... | | 著者 | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | 登録日 | 2023-10-17 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-08-14 | | 実験手法 | X-RAY DIFFRACTION (1.925 Å) | | 主引用文献 | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z203039992
To Be Published
|
|
6XG4
 
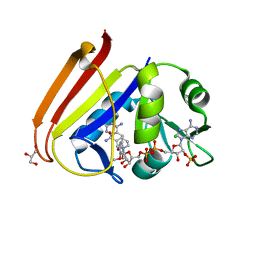 | | X-ray structure of Escherichia coli dihydrofolate reductase L28R mutant in complex with trimethoprim | | 分子名称: | CHLORIDE ION, Dihydrofolate reductase, GLYCEROL, ... | | 著者 | Gaszek, I.K, Manna, M.S, Borek, D, Toprak, E. | | 登録日 | 2020-06-16 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A trimethoprim derivative impedes antibiotic resistance evolution.
Nat Commun, 12, 2021
|
|
6XG5
 
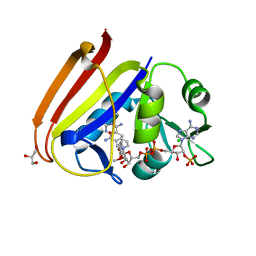 | | X-ray structure of Escherichia coli dihydrofolate reductase in complex with trimethoprim | | 分子名称: | CHLORIDE ION, Dihydrofolate reductase, GLYCEROL, ... | | 著者 | Gaszek, I.K, Manna, M.S, Borek, D, Toprak, E. | | 登録日 | 2020-06-16 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A trimethoprim derivative impedes antibiotic resistance evolution.
Nat Commun, 12, 2021
|
|
8ULM
 
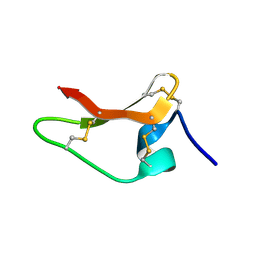 | |
6E29
 
 | |
8U26
 
 | |
8U2C
 
 | |
6EK6
 
 | | Crystal structure of KDM5B in complex with S49195a. | | 分子名称: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Lysine-specific demethylase 5B,Lysine-specific demethylase 5B, ... | | 著者 | Srikannathasan, V, Szykowska, A, Newman, J.A, Ruda, G.F, Strain-Damerell, C, Burgess-Brown, N.A, Vazquez-Rodriguez, S, Wright, M, Brennan, P.E, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Huber, K, von Delft, F. | | 登録日 | 2017-09-25 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal structure of KDM5B in complex with S49195a.
To be published
|
|
8U28
 
 | |
8UGQ
 
 | |
1TVM
 
 | | NMR structure of enzyme GatB of the galactitol-specific phosphoenolpyruvate-dependent phosphotransferase system | | 分子名称: | PTS system, galactitol-specific IIB component | | 著者 | Volpon, L, Young, C.R, Lim, N.S, Iannuzzi, P, Cygler, M, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | 登録日 | 2004-06-29 | | 公開日 | 2005-09-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the enzyme GatB of the galactitol-specific phosphoenolpyruvate-dependent phosphotransferase system and its interaction with GatA.
Protein Sci., 15, 2006
|
|
6E8L
 
 | | Crystal Structure of Alkyl hydroperoxidase D (AhpD) from Streptococcus pneumoniae (Strain D39/ NCTC 7466) | | 分子名称: | Alkyl hydroperoxide reductase AhpD | | 著者 | Meng, Y, Davies, J, North, R, Coombes, D, Horne, C, Hampton, M, Dobson, R. | | 登録日 | 2018-07-30 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-function analyses of alkylhydroperoxidase D fromStreptococcus pneumoniaereveal an unusual three-cysteine active site architecture.
J.Biol.Chem., 295, 2020
|
|
3AQF
 
 | | Crystal structure of the human CRLR/RAMP2 extracellular complex | | 分子名称: | Calcitonin gene-related peptide type 1 receptor, Receptor activity-modifying protein 2 | | 著者 | Kusano, S, Kukimono-Niino, M, Shirouzu, M, Shindo, T, Yokoyama, S. | | 登録日 | 2010-10-29 | | 公開日 | 2011-11-02 | | 最終更新日 | 2012-07-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for extracellular interactions between calcitonin receptor-like receptor and receptor activity-modifying protein 2 for adrenomedullin-specific binding
Protein Sci., 21, 2012
|
|
8V1P
 
 | | CRYSTAL STRUCTURE OF GID4 IN COMPLEX WITH UBF9092 | | 分子名称: | Glucose-induced degradation protein 4 homolog, N,N~2~-bis[(4-methoxyphenyl)methyl]glycinamide | | 著者 | Dong, C, Dong, A, Calabrese, M, Wang, F, Owen, D, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2023-11-21 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF GID4 IN COMPLEX WITH UBF9092
To be published
|
|
8QHM
 
 | |
7V7M
 
 | | crystal structure of SARS-CoV-2 3CL protease | | 分子名称: | 3C-like proteinase | | 著者 | Yi, Y, Zhang, M, Ye, M. | | 登録日 | 2021-08-21 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Schaftoside inhibits 3CL pro and PL pro of SARS-CoV-2 virus and regulates immune response and inflammation of host cells for the treatment of COVID-19.
Acta Pharm Sin B, 12, 2022
|
|
