2Y8N
 
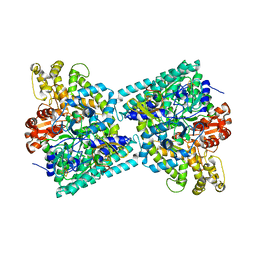 | | Crystal structure of glycyl radical enzyme | | 分子名称: | 4-HYDROXYPHENYLACETATE DECARBOXYLASE LARGE SUBUNIT, 4-HYDROXYPHENYLACETATE DECARBOXYLASE SMALL SUBUNIT, IRON/SULFUR CLUSTER | | 著者 | Martins, B.M, Blaser, M, Feliks, M, Ullmann, G.M, Selmer, T. | | 登録日 | 2011-02-08 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Basis for a Kolbe-Type Decarboxylation Catalyzed by a Glycyl Radical Enzyme.
J.Am.Chem.Soc., 133, 2011
|
|
5JDV
 
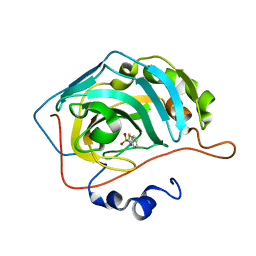 | | Human carbonic anhydrase II (F131W) complexed with benzo[d]thiazole-2-sulfonamide | | 分子名称: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | 著者 | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | 登録日 | 2016-04-17 | | 公開日 | 2017-01-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5ESA
 
 | |
5JLC
 
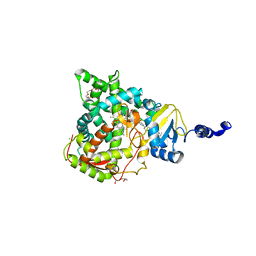 | | Structure of CYP51 from the pathogen Candida glabrata | | 分子名称: | 2-[(2R)-butan-2-yl]-4-{4-[4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazin-1-yl]phenyl}-2,4-dihydro-3H-1,2,4-triazol-3-one, CHLORIDE ION, Lanosterol 14-alpha demethylase, ... | | 著者 | Keniya, M.V, Sabherwal, M, Wilson, R.K, Sagatova, A.A, Tyndall, J.D.A, Monk, B.C. | | 登録日 | 2016-04-26 | | 公開日 | 2016-05-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structures of Full-Length Lanosterol 14 alpha-Demethylases of Prominent Fungal Pathogens Candida albicans and Candida glabrata Provide Tools for Antifungal Discovery.
Antimicrob.Agents Chemother., 62, 2018
|
|
8P67
 
 | | Crystal structure of Thermothelomyces thermophila (double mutant EE) in complex with aldotetrauronic acid | | 分子名称: | 1,2-ETHANEDIOL, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, GH30 family xylanase, ... | | 著者 | Dimarogona, M, Pentari, C, Kosinas, C, Topakas, E. | | 登録日 | 2023-05-25 | | 公開日 | 2024-05-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Structural and molecular insights into a bifunctional glycoside hydrolase 30 xylanase specific to glucuronoxylan.
Biotechnol.Bioeng., 121, 2024
|
|
5F1Q
 
 | | Crystal Structure of Periplasmic Dipeptide Transport Protein from Yersinia pestis | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kim, Y, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-11-30 | | 公開日 | 2015-12-23 | | 最終更新日 | 2019-11-27 | | 実験手法 | X-RAY DIFFRACTION (1.956 Å) | | 主引用文献 | Crystal Structure of Periplasmic Dipeptide Transport Protein from Yersinia pestis
To Be Published
|
|
8Q0N
 
 | | HACE1 in complex with RAC1 Q61L | | 分子名称: | E3 ubiquitin-protein ligase HACE1, GUANOSINE-5'-TRIPHOSPHATE, Ras-related C3 botulinum toxin substrate 1, ... | | 著者 | Wolter, M, Duering, J, Dienemann, C, Lorenz, S. | | 登録日 | 2023-07-28 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural mechanisms of autoinhibition and substrate recognition by the ubiquitin ligase HACE1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3WOL
 
 | | Crystal structure of the DAP BII dipeptide complex I | | 分子名称: | GLYCEROL, TYROSINE, VALINE, ... | | 著者 | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | 登録日 | 2013-12-29 | | 公開日 | 2014-09-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
8PWL
 
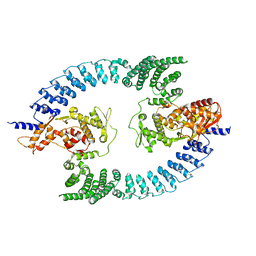 | |
6ZGK
 
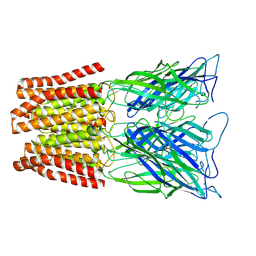 | | GLIC pentameric ligand-gated ion channel, pH 3 | | 分子名称: | Proton-gated ion channel | | 著者 | Rovsnik, U, Zhuang, Y, Forsberg, B.O, Carroni, M, Yvonnesdotter, L, Howard, R.J, Lindahl, E. | | 登録日 | 2020-06-18 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Dynamic closed states of a ligand-gated ion channel captured by cryo-EM and simulations.
Life Sci Alliance, 4, 2021
|
|
5ESL
 
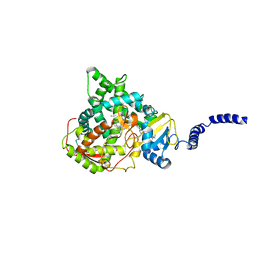 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) T322I mutant complexed with itraconazole | | 分子名称: | 2-[(2R)-butan-2-yl]-4-{4-[4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazin-1-yl]phenyl}-2,4-dihydro-3H-1,2,4-triazol-3-one, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Sagatova, A, Keniya, M.V, Wilson, R.K, Sabherwal, M, Tyndall, J.D.A, Monk, B.C. | | 登録日 | 2015-11-16 | | 公開日 | 2016-11-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structures of lanosterol 14-alpha demethylase mutants.
To Be Published
|
|
2YUY
 
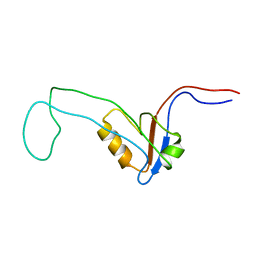 | | Solution Structure of PDZ domain of Rho GTPase Activating Protein 21 | | 分子名称: | Rho GTPase activating protein 21 | | 著者 | Niraula, T.N, Yoneyama, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-04-06 | | 公開日 | 2008-04-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of PDZ domain of Rho GTPase Activating Protein 21
To be Published
|
|
7TAM
 
 | |
8PWR
 
 | | TINA-conjugated antiparallel DNA triplex | | 分子名称: | DNA (5'-D(*AP*GP*GP*AP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*TP*CP*CP*T)-3'), DNA (5'-D(*TP*GP*GP*TP*GP*(J32)P*GP*T)-3') | | 著者 | Garavis, M, Edwards, P.J.B, Serrano-Chacon, I, Doluca, O, Filichev, V.V, Gonzalez, C. | | 登録日 | 2023-07-21 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Understanding intercalative modulation of G-rich sequence folding: solution structure of a TINA-conjugated antiparallel DNA triplex.
Nucleic Acids Res., 52, 2024
|
|
5ESN
 
 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) T322I mutant structure | | 分子名称: | Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Sagatova, A, Keniya, M.V, Wilson, R.K, Sabherwal, M, Tyndall, J.D.A, Monk, B.C. | | 登録日 | 2015-11-16 | | 公開日 | 2016-11-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structures of lanosterol 14-alpha demethylase mutants.
To Be Published
|
|
2YE0
 
 | | X-ray structure of the cyan fluorescent protein mTurquoise (K206A mutant) | | 分子名称: | GREEN FLUORESCENT PROTEIN | | 著者 | von Stetten, D, Goedhart, J, Noirclerc-Savoye, M, Lelimousin, M, Joosen, L, Hink, M.A, van Weeren, L, Gadella, T.W.J, Royant, A. | | 登録日 | 2011-03-25 | | 公開日 | 2012-03-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Structure-Guided Evolution of Cyan Fluorescent Proteins Towards a Quantum Yield of 93%
Nat.Commun, 3, 2012
|
|
5ESM
 
 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) T322I mutant complexed with fluconazole | | 分子名称: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Sagatova, A, Keniya, M.V, Wilson, R.K, Sabherwal, M, Tyndall, J.D.A, Monk, B.C. | | 登録日 | 2015-11-16 | | 公開日 | 2016-11-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of lanosterol 14-alpha demethylase mutants.
To Be Published
|
|
3WON
 
 | | Crystal structure of the DAP BII dipeptide complex III | | 分子名称: | GLYCEROL, TYROSINE, VALINE, ... | | 著者 | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | 登録日 | 2013-12-29 | | 公開日 | 2014-09-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
6ZGD
 
 | | GLIC pentameric ligand-gated ion channel, pH 7 | | 分子名称: | Proton-gated ion channel | | 著者 | Rovsnik, U, Zhuang, Y, Forsberg, B.O, Carroni, M, Yvonnesdotter, L, Howard, R.J, Lindahl, E. | | 登録日 | 2020-06-18 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Dynamic closed states of a ligand-gated ion channel captured by cryo-EM and simulations.
Life Sci Alliance, 4, 2021
|
|
6ZGJ
 
 | | GLIC pentameric ligand-gated ion channel, pH 5 | | 分子名称: | Proton-gated ion channel | | 著者 | Rovsnik, U, Zhuang, Y, Forsberg, B.O, Carroni, M, Yvonnesdotter, L, Howard, R.J, Lindahl, E. | | 登録日 | 2020-06-18 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Dynamic closed states of a ligand-gated ion channel captured by cryo-EM and simulations.
Life Sci Alliance, 4, 2021
|
|
5EL4
 
 | | Structure of T. thermophilus 70S ribosome complex with mRNA and tRNALys in the A-site with a U-U mismatch in the first position | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Rozov, A, Demeshkina, N, Khusainov, I, Yusupov, M, Yusupova, G. | | 登録日 | 2015-11-04 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code.
Nat Commun, 7, 2016
|
|
8PV8
 
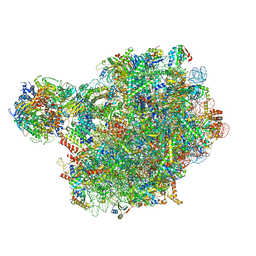 | | Chaetomium thermophilum pre-60S State 4 - post-5S rotation with Rix1 complex without Foot - composite structure | | 分子名称: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | 登録日 | 2023-07-17 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
5EVH
 
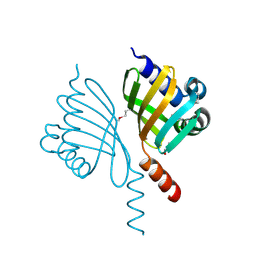 | | Crystal structure of known function protein from Kribbella flavida DSM 17836 | | 分子名称: | GLYCEROL, Uncharacterized protein | | 著者 | Chang, C, Duke, N, Endres, M, Chhor, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-11-19 | | 公開日 | 2015-12-02 | | 実験手法 | X-RAY DIFFRACTION (1.852 Å) | | 主引用文献 | Crystal structure of known function protein from Kribbella flavida DSM 17836
To Be Published
|
|
5J8J
 
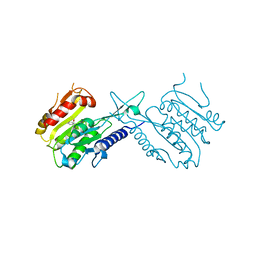 | |
2YNV
 
 | | Cys221 oxidized, Mono zinc GIM-1 - GIM-1-Ox. Crystal structures of Pseudomonas aeruginosa GIM-1: active site plasticity in metallo-beta- lactamases | | 分子名称: | GIM-1 PROTEIN, MAGNESIUM ION, ZINC ION | | 著者 | Borra, P.S, Samuelsen, O, Spencer, J, Lorentzen, M.S, Leiros, H.-K.S. | | 登録日 | 2012-10-18 | | 公開日 | 2013-07-24 | | 最終更新日 | 2018-06-20 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal Structures of Pseudomonas Aeruginosa Gim-1: Active-Site Plasticity in Metallo-Beta-Lactamases.
Antimicrob.Agents Chemother., 57, 2013
|
|
