6G87
 
 | | Flavonoid-responsive Regulator FrrA | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, TetR/AcrR family transcriptional regulator | | Authors: | Werner, N, Hoppen, J, Palm, G, Werten, S, Goettfert, M, Hinrichs, W. | | Deposit date: | 2018-04-07 | | Release date: | 2019-04-24 | | Last modified: | 2021-08-18 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | The induction mechanism of the flavonoid-responsive regulator FrrA.
Febs J., 2021
|
|
3OBG
 
 | |
6K0I
 
 | | Crystal Structure of UDP-glucose 4-epimerase from Bifidobacterium longum in complex with NAD+ and UDP-Glc | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Nam, Y.-W, Nishimoto, M, Arakawa, T, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2019-05-06 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for broad substrate specificity of UDP-glucose 4-epimerase in the human milk oligosaccharide catabolic pathway of Bifidobacterium longum.
Sci Rep, 9, 2019
|
|
1TOF
 
 | | THIOREDOXIN H (OXIDIZED FORM), NMR, 23 STRUCTURES | | Descriptor: | THIOREDOXIN H | | Authors: | Mittard, V, Blackledge, M.J, Stein, M, Jacquot, J.-P, Marion, D, Lancelin, J.-M. | | Deposit date: | 1996-05-30 | | Release date: | 1996-12-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oxidised thioredoxin h from the eukaryotic green alga Chlamydomonas reinhardtii.
Eur.J.Biochem., 243, 1997
|
|
4LC1
 
 | | MeaB, A Bacterial Homolog of MMAA, Bound to GDP and crystallized in the presence of GDP and [AlF4]- | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, Methylmalonyl-CoA mutase accessory protein | | Authors: | Koutmos, M, Padovani, D, Lofgren, M, Banerjee, R. | | Deposit date: | 2013-06-21 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Autoinhibition and Signaling by the Switch II Motif in the G-protein Chaperone of a Radical B12 Enzyme.
J.Biol.Chem., 288, 2013
|
|
5A7T
 
 | | Crystal structure of Sulfolobus acidocaldarius Trm10 at 2.4 angstrom resolution. | | Descriptor: | DI(HYDROXYETHYL)ETHER, TRNA (ADENINE(9)-N1)-METHYLTRANSFERASE | | Authors: | Van Laer, B, Roovers, M, Wauters, L, Kasprzak, J, Dyzma, M, Deyaert, E, Feller, A, Bujnicki, J, Droogmans, L, Versees, W. | | Deposit date: | 2015-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Insights Into tRNA Binding and Adenosine N1-Methylation by an Archaeal Trm10 Homologue.
Nucleic Acids Res., 44, 2016
|
|
2OXT
 
 | | Crystal structure of Meaban virus nucleoside-2'-O-methyltransferase | | Descriptor: | NUCLEOSIDE-2'-O-METHYLTRANSFERASE, S-ADENOSYLMETHIONINE | | Authors: | Mastrangelo, E, Milani, M, Bollati, M, Bolognesi, M. | | Deposit date: | 2007-02-21 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural bases for substrate recognition and activity in Meaban virus nucleoside-2'-O-methyltransferase
Protein Sci., 16, 2007
|
|
6CWV
 
 | | Protein Tyrosine Phosphatase 1B A122S mutant | | Descriptor: | MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Hjortness, M, Zwart, P, Sankaran, B, Fox, J.M. | | Deposit date: | 2018-03-31 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.98002291 Å) | | Cite: | Evolutionarily Conserved Allosteric Communication in Protein Tyrosine Phosphatases.
Biochemistry, 57, 2018
|
|
1KZ6
 
 | | Mutant enzyme W63Y/L119F Lumazine Synthase from S.pombe | | Descriptor: | 6,7-Dimethyl-8-ribityllumazine Synthase, PHOSPHATE ION | | Authors: | Gerhardt, S, Haase, I, Steinbacher, S, Kaiser, J.T, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase.
J.Mol.Biol., 318, 2002
|
|
1BZQ
 
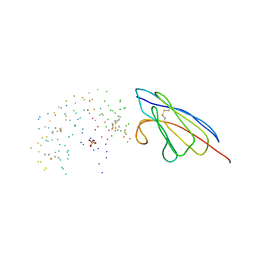 | | COMPLEX OF A DROMEDARY SINGLE-DOMAIN VHH ANTIBODY FRAGMENT WITH RNASE A | | Descriptor: | PHOSPHATE ION, PROTEIN (ANTIBODY CAB-RN05), PROTEIN (RNASE A) | | Authors: | Decanniere, K, Desmyter, A, Gahroudhi, M, Lauwereys, M, Muyldermans, S, Wyns, L. | | Deposit date: | 1998-11-03 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A single-domain antibody fragment in complex with RNase A: non-canonical loop structures and nanomolar affinity using two CDR loops.
Structure Fold.Des., 7, 1999
|
|
1Y25
 
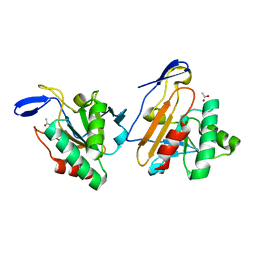 | | structure of mycobacterial thiol peroxidase Tpx | | Descriptor: | ACETATE ION, Probable thiol peroxidase | | Authors: | Stehr, M, Hoffmann, B, Jger, T, Singh, M, Hecht, H.J. | | Deposit date: | 2004-11-20 | | Release date: | 2005-11-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the inactive variant C60S of Mycobacterium tuberculosis thiol peroxidase
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3ODI
 
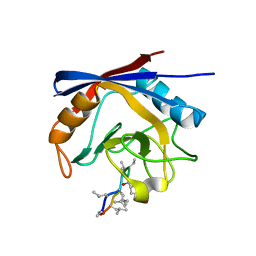 | | Crystal structure of cyclophilin A in complex with Voclosporin E-ISA247 | | Descriptor: | Cyclophilin A, Voclosporin | | Authors: | Kuglstatter, A, Stihle, M, Benz, J, Hennig, M. | | Deposit date: | 2010-08-11 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the cyclophilin A binding affinity and immunosuppressive potency of E-ISA247 (voclosporin).
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4LJM
 
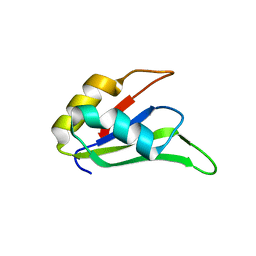 | |
3ODL
 
 | | Crystal structure of cyclophilin A in complex with Voclosporin Z-ISA247 | | Descriptor: | Cyclophilin A, Voclosporin | | Authors: | Kuglstatter, A, Stihle, M, Benz, J, Hennig, M. | | Deposit date: | 2010-08-11 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis for the cyclophilin A binding affinity and immunosuppressive potency of E-ISA247 (voclosporin).
Acta Crystallogr.,Sect.D, 67, 2011
|
|
6GD3
 
 | | Structure of HuR RRM3 in complex with RNA (UAUUUA) | | Descriptor: | ELAV-like protein 1, RNA (5'-R(P*UP*AP*UP*UP*UP*A)-3'), SODIUM ION | | Authors: | Pabis, M, Sattler, M. | | Deposit date: | 2018-04-21 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs.
Nucleic Acids Res., 47, 2019
|
|
6JT5
 
 | | Crystal structure of PQQ doamin of Pyranose Dehydrogenase from Coprinopsis cinerea: apo-from | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular PQQ-dependent sugar dehydrogenase, ... | | Authors: | Takeda, K, Ishida, T, Yoshida, M, Samejima, M, Ohno, H, Igarashi, K, Nakamura, N. | | Deposit date: | 2019-04-09 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the Catalytic and CytochromebDomains in a Eukaryotic Pyrroloquinoline Quinone-Dependent Dehydrogenase.
Appl.Environ.Microbiol., 85, 2019
|
|
5CGA
 
 | | Structure of Hydroxyethylthiazole kinase ThiM from Staphylococcus aureus in complex with substrate analog 2-(1,3,5-trimethyl-1H-pyrazole-4-yl)ethanol | | Descriptor: | 2-(1,3,5-trimethyl-1H-pyrazol-4-yl)ethanol, Hydroxyethylthiazole kinase, MAGNESIUM ION | | Authors: | Kuenz, M, Drebes, J, Windshuegel, B, Cang, H, Wrenger, C, Betzel, C. | | Deposit date: | 2015-07-09 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of ThiM from Vitamin B1 biosynthetic pathway of Staphylococcus aureus - Insights into a novel pro-drug approach addressing MRSA infections.
Sci Rep, 6, 2016
|
|
8AZS
 
 | | Type I amyloid-beta 42 filaments from high-spin supernatants of aqueous extracts from Alzheimer's disease brains | ABeta42 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Yang, Y, Stern, M.A, Meunier, L.A, Liu, W, Cai, Y.Q, Ericsson, M, Liu, L, Selkoe, J.D, Goedert, M, Scheres, H.W.S. | | Deposit date: | 2022-09-06 | | Release date: | 2022-11-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Abundant A beta fibrils in ultracentrifugal supernatants of aqueous extracts from Alzheimer's disease brains.
Neuron, 111, 2023
|
|
1L7E
 
 | | Crystal Structure of R. rubrum Transhydrogenase Domain I with Bound NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, nicotinamide nucleotide Transhydrogenase, subunit alpha 1 | | Authors: | Prasad, G.S, Wahlberg, M, Sridhar, V, Yamaguchi, M, Hatefi, Y, Stout, C.D. | | Deposit date: | 2002-03-14 | | Release date: | 2002-11-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Transhydrogenase Domain I
with and without Bound NADH
Biochemistry, 41, 2002
|
|
3OBJ
 
 | |
3OFW
 
 | | Crystal structure of recombinant Kunitz Type serine protease Inhibitor-1 from the Carribean sea anemone stichodactyla helianthus | | Descriptor: | CHLORIDE ION, Kunitz-type proteinase inhibitor SHPI-1 | | Authors: | Garcia-Fernandez, R, Redecke, L, Pons, T, Perbandt, M, Talavera, A, Gil, D, Gonzalez, Y, de los Angeles Chavez, M, Betzel, C. | | Deposit date: | 2010-08-16 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the recombinant BPTI/Kunitz-type inhibitor rShPI-1A from the marine invertebrate Stichodactyla helianthus.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
6I0J
 
 | | Crystal structure of human carbonic anhydrase I in complex with the 4-({[4-chloro-3-(trifluoromethyl)phenyl]carbamoyl}amino)phenyl sulfamate inhibitor | | Descriptor: | ACETATE ION, Carbonic anhydrase 1, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C.T, Bozdag, M, Chiapponi, D. | | Deposit date: | 2018-10-26 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Carbonic anhydrase inhibitors based on sorafenib scaffold: Design, synthesis, crystallographic investigation and effects on primary breast cancer cells.
Eur.J.Med.Chem., 182, 2019
|
|
5UJH
 
 | | ov-GRN12-34 | | Descriptor: | Granulin | | Authors: | Bansal, P, Smout, M, Wilson, D, Caceres, C.C, Dastpeyman, M, Sotillo, J, Seifert, J, Brindley, P, Loukas, A, Daly, N. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Development of a Potent Wound Healing Agent Based on the Liver Fluke Granulin Structural Fold.
J. Med. Chem., 60, 2017
|
|
1TUI
 
 | | INTACT ELONGATION FACTOR TU IN COMPLEX WITH GDP | | Descriptor: | ELONGATION FACTOR TU, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Polekhina, G, Thirup, S, Kjeldgaard, M, Nissen, P, Lippmann, C, Nyborg, J. | | Deposit date: | 1996-05-23 | | Release date: | 1997-06-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Helix unwinding in the effector region of elongation factor EF-Tu-GDP.
Structure, 4, 1996
|
|
1LBQ
 
 | | The crystal structure of Saccharomyces cerevisiae ferrochelatase | | Descriptor: | Ferrochelatase | | Authors: | Karlberg, T, Lecerof, D, Gora, M, Silvegren, G, Labbe-Bois, R, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 2002-04-04 | | Release date: | 2002-11-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Metal binding to Saccharomyces cerevisiae ferrochelatase
Biochemistry, 41, 2002
|
|
