5H24
 
 | | EED in complex with PRC2 allosteric inhibitor compound 8 | | Descriptor: | 5-(furan-2-ylmethylamino)-[1,2,4]triazolo[4,3-a]pyridine-6-carbonitrile, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-14 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of First-in-Class, Potent, and Orally Bioavailable Embryonic Ectoderm Development (EED) Inhibitor with Robust Anticancer Efficacy
J. Med. Chem., 60, 2017
|
|
6T3V
 
 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - malic acid | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Rutkiewicz, M, Bujacz, A, Rum, J, Bujacz, G. | | Deposit date: | 2019-10-11 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials, 14, 2021
|
|
5GWR
 
 | | 4-hydroxyisoleucine dehydrogenase complexed with NADH | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxyisolecuine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Shi, X, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2016-09-13 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Engineering a short-chain dehydrogenase/reductase for the stereoselective production of (2S,3R,4S)-4-hydroxyisoleucine with three asymmetric centers
Sci Rep, 7, 2017
|
|
6T5E
 
 | | Hydroxylamine Oxidoreductase from Brocadia fulgida | | Descriptor: | HEME C, Hydroxylamine oxidoreductase-like protein, SULFATE ION | | Authors: | Akram, M, Dietl, A, Mueller, M, Barends, T. | | Deposit date: | 2019-10-16 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Purification of the key enzyme complexes of the anammox pathway from DEMON sludge.
Biopolymers, 112, 2021
|
|
1KXV
 
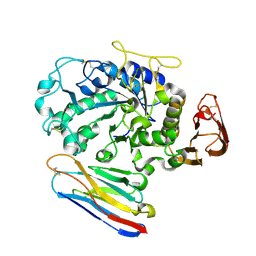 | | Camelid VHH Domains in Complex with Porcine Pancreatic alpha-Amylase | | Descriptor: | ALPHA-AMYLASE, PANCREATIC, CAMELID VHH DOMAIN CAB10 | | Authors: | Desmyter, A, Spinelli, S, Payan, F, Lauwereys, M, Wyns, L, Muyldermans, S, Cambillau, C. | | Deposit date: | 2002-02-01 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology.
J.Biol.Chem., 277, 2002
|
|
6HAY
 
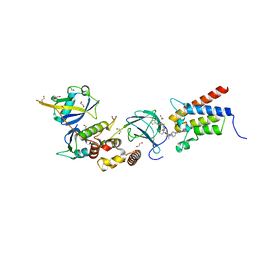 | | Crystal structure of PROTAC 1 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[[2-[2-[2-[2-[4-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazin-1-yl]ethoxy]ethoxy]ethoxy]-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Roy, M, Bader, G, Diers, E, Trainor, N, Farnaby, W, Ciulli, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design.
Nat.Chem.Biol., 15, 2019
|
|
3NZX
 
 | | Crystal structure of the yeast 20S proteasome in complex with ligand 2c | | Descriptor: | Proteasome component C1, Proteasome component C11, Proteasome component C5, ... | | Authors: | Groll, M, Gallastegui, N, Marechal, X, Le Ravalec, V, Basse, N, Richy, N, Genin, E, Huber, R, Moroder, M, Vidal, V, Reboud-Ravaux, M. | | Deposit date: | 2010-07-17 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 20S proteasome inhibition: designing noncovalent linear peptide mimics of the natural product TMC-95A.
Chemmedchem, 5, 2010
|
|
3OFK
 
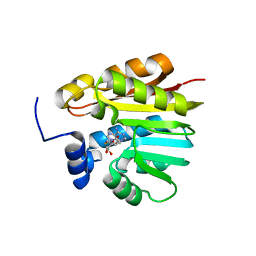 | | Crystal structure of N-methyltransferase NodS from Bradyrhizobium japonicum WM9 in complex with S-adenosyl-l-homocysteine (SAH) | | Descriptor: | Nodulation protein S, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Cakici, O, Sikorski, M, Stepkowski, T, Bujacz, G, Jaskolski, M. | | Deposit date: | 2010-08-15 | | Release date: | 2010-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of NodS N-Methyltransferase from Bradyrhizobium japonicum in Ligand-Free Form and as SAH Complex.
J.Mol.Biol., 404, 2010
|
|
5T09
 
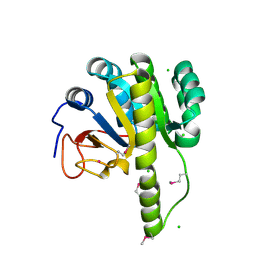 | | The structure of the type III effector HopBA1 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, Type III secretion system effector HopBA1 | | Authors: | Cherkis, K, Machius, M, Nishimura, M.T, Dangl, J.L. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | TIR-only protein RBA1 recognizes a pathogen effector to regulate cell death in Arabidopsis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
12GS
 
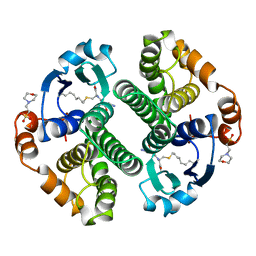 | | GLUTATHIONE S-TRANSFERASE COMPLEXED WITH S-NONYL-GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE, L-gamma-glutamyl-S-nonyl-L-cysteinylglycine | | Authors: | Oakley, A.J, Lo Bello, M, Parker, M.W. | | Deposit date: | 1997-11-19 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Ligandin (Non-Substrate) Binding Site of Human Pi Class Glutathione Transferase is Located in the Electrophile Binding Site (H-Site).
J.Mol.Biol., 291, 1999
|
|
13GS
 
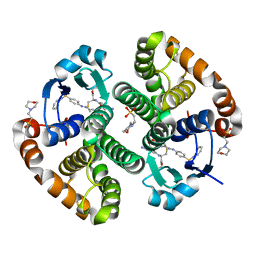 | | GLUTATHIONE S-TRANSFERASE COMPLEXED WITH SULFASALAZINE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-HYDROXY-(5-([4-(2-PYRIDINYLAMINO)SULFONYL]PHENYL)AZO)BENZOIC ACID, GLUTATHIONE, ... | | Authors: | Oakley, A.J, Lo Bello, M, Parker, M.W. | | Deposit date: | 1997-11-20 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Ligandin (Non-Substrate) Binding Site of Human Pi Class Glutathione Transferase is Located in the Electrophile Binding Site (H-Site).
J.Mol.Biol., 291, 1999
|
|
6FKY
 
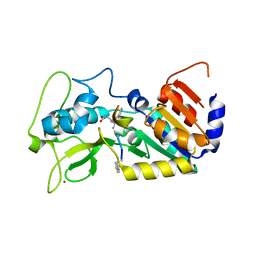 | |
5IJC
 
 | | The crystal structure of mouse TLR4/MD-2/neoseptin-3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6FKG
 
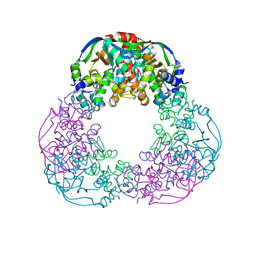 | | Crystal structure of the M.tuberculosis MbcT-MbcA toxin-antitoxin complex. | | Descriptor: | GLYCEROL, Rv1989c (MbcT), Rv1990c (MbcA) | | Authors: | Freire, D.M, Cianci, M, Pogenberg, V, Schneider, T.R, Wilmanns, M, Parret, A.H.A. | | Deposit date: | 2018-01-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An NAD+Phosphorylase Toxin Triggers Mycobacterium tuberculosis Cell Death.
Mol.Cell, 73, 2019
|
|
1X9U
 
 | | Umecyanin from Horse Raddish- Crystal Structure of the reduced form | | Descriptor: | COPPER (II) ION, Umecyanin | | Authors: | Koch, M, Velarde, M, Harrison, M.D, Echt, S, Fischer, M, Messerschmidt, A, Dennison, C. | | Deposit date: | 2004-08-24 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Oxidized and Reduced Stellacyanin from Horseradish Roots
J.Am.Chem.Soc., 127, 2005
|
|
6VNZ
 
 | |
1KSH
 
 | | Complex of Arl2 and PDE delta, Crystal Form 2 (native) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Hanzal-Bayer, M, Renault, L, Roversi, P, Wittinghofer, A, Hillig, R.C. | | Deposit date: | 2002-01-13 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The complex of Arl2-GTP and PDE delta: from structure to function.
EMBO J., 21, 2002
|
|
6HC3
 
 | | TFAM bound to Site-X | | Descriptor: | DNA (5'-D(*TP*TP*TP*GP*GP*TP*GP*GP*AP*AP*AP*TP*TP*TP*TP*TP*TP*GP*TP*TP*AP*G)-3'), DNA/RNA (5'-D(*TP*AP*AP*CP*AP*AP*AP*AP*AP*AP*TP*TP*TP*CP*CP*AP*CP*CP*AP*AP*AP*C)-3'), L(+)-TARTARIC ACID, ... | | Authors: | Fernandez-Millan, P, Cuppari, A, Tarres-Sole, A, Rubio-Cosials, A, Lyonnais, S, Sola, M. | | Deposit date: | 2018-08-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | DNA specificities modulate the binding of human transcription factor A to mitochondrial DNA control region.
Nucleic Acids Res., 47, 2019
|
|
5FGH
 
 | |
1KLV
 
 | | Solution Structure and Backbone Dynamics of GABARAP, GABAA Receptor associated protein | | Descriptor: | GABA(A) Receptor associated protein | | Authors: | Kouno, T, Miura, K, Tada, M, Kanematsu, T, Tate, S, Shirakawa, M, Hirata, M, Kawano, K. | | Deposit date: | 2001-12-13 | | Release date: | 2003-10-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and '5N resonance assignments of GABARAP, GABAA receptor associated protein.
J.Biomol.Nmr, 22, 2002
|
|
8A8L
 
 | | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with a heme-coordinated fragment | | Descriptor: | 6-methoxy-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole, Cytochrome P450 protein, GLYCEROL, ... | | Authors: | Snee, M, Katariya, M, Levy, C. | | Deposit date: | 2022-06-23 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with a heme-coordinated fragment
To Be Published
|
|
1CB2
 
 | | CELLOBIOHYDROLASE II, CATALYTIC DOMAIN, MUTANT Y169F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE II, alpha-D-mannopyranose | | Authors: | Kleywegt, G.J, Szardenings, M, Jones, T.A. | | Deposit date: | 1995-11-25 | | Release date: | 1996-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site of Trichoderma reesei cellobiohydrolase II: the role of tyrosine 169.
Protein Eng., 9, 1996
|
|
6I8X
 
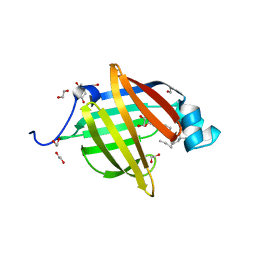 | | As-p18, an extracellular fatty acid binding protein | | Descriptor: | 1,2-ETHANEDIOL, Fatty acid-binding protein homolog, TRIS(HYDROXYETHYL)AMINOMETHANE, ... | | Authors: | Gabrielsen, M, Riboldi-Tunnicliffe, A, Ibanez-Shimabukuro, M, Smith, B.O. | | Deposit date: | 2018-11-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | As-p18, an extracellular fatty acid binding protein of nematodes, exhibits unusual structural features.
Biosci.Rep., 2019
|
|
3MLE
 
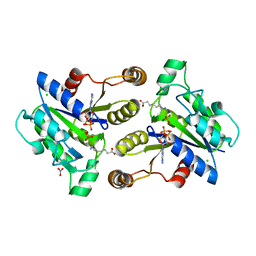 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori cocrystallized with ATP | | Descriptor: | 8-aminooctanoic acid, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Nicholls, R, Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Joachimiak, A, Murshudov, G, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-16 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
5FSC
 
 | | Structure of tectonin 2 from Laccaria bicolor in complex with allyl- alpha_4-methyl-mannoside | | Descriptor: | MAGNESIUM ION, TECTONIN 2, prop-2-en-1-yl 4-O-methyl-alpha-D-mannopyranoside | | Authors: | Sommer, R, Bleuer, S, Titz, A, Kunzler, M, Varrot, A. | | Deposit date: | 2016-01-04 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of Fungal Tectonin in Complex with O-Methylated Glycans Suggest Key Role in Innate Immune Defense.
Structure, 26, 2018
|
|
