8R7H
 
 | | Cryo-EM structure of Human SHMT1 | | Descriptor: | Serine hydroxymethyltransferase, cytosolic | | Authors: | Spizzichino, S, Marabelli, C, Bharadwaj, A, Jakobi, A.J, Chaves-Sanjuan, A, Giardina, G, Bolognesi, M, Cutruzzola, F. | | Deposit date: | 2023-11-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structure-based mechanism of riboregulation of the metabolic enzyme SHMT1.
Mol.Cell, 84, 2024
|
|
8HIZ
 
 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | Descriptor: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | Deposit date: | 2022-11-22 | | Release date: | 2023-02-08 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
4ORS
 
 | | Three-dimensional structure of the C65A mutant of Human lipocalin-type Prostaglandin D Synthase apo-form | | Descriptor: | Prostaglandin-H2 D-isomerase, SULFATE ION | | Authors: | Perduca, M, Bovi, M, Bertinelli, M, Bertini, E, Destefanis, L, Carrizo, M.E, Capaldi, S, Monaco, H.L. | | Deposit date: | 2014-02-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution structures of mutants of residues that affect access to the ligand-binding cavity of human lipocalin-type prostaglandin D synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1HSZ
 
 | | HUMAN BETA-1 ALCOHOL DEHYDROGENASE (ADH1B*1) | | Descriptor: | CLASS I ALCOHOL DEHYDROGENASE 1, BETA SUBUNIT, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Niederhut, M.S, Gibbons, B.J, Perez-Miller, S, Hurley, T.D. | | Deposit date: | 2000-12-27 | | Release date: | 2001-01-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of the three human class I alcohol dehydrogenases.
Protein Sci., 10, 2001
|
|
8OKR
 
 | |
3BRI
 
 | | Crystal Structure of apo-LC8 | | Descriptor: | ACETATE ION, Dynein light chain 1, cytoplasmic, ... | | Authors: | Benison, G, Karplus, P.A, Barbar, E, Chiodo, M. | | Deposit date: | 2007-12-21 | | Release date: | 2008-12-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Interplay of Ligand Binding and Quaternary Structure in the Diverse Interactions of Dynein Light Chain LC8.
J.Mol.Biol., 384, 2008
|
|
5X9G
 
 | |
3C1C
 
 | | The effect of H3 K79 dimethylation and H4 K20 trimethylation on nucleosome and chromatin structure | | Descriptor: | Histone 2, H2bf, Histone H2A type 1, ... | | Authors: | Lu, X, Simon, M, Chodaparambil, J, Hansen, J, Shokat, K, Luger, K. | | Deposit date: | 2008-01-22 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure.
Nat.Struct.Mol.Biol., 15, 2008
|
|
1QXS
 
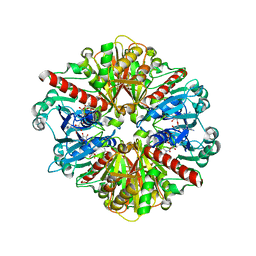 | | CRYSTAL STRUCTURE OF Trypanosoma cruzi GLYCERALDEHYDE-3- PHOSPHATE DEHYDROGENASE COMPLEXED WITH AN ANALOGUE OF 1,3- BisPHOSPHO-D-GLYCERIC ACID | | Descriptor: | 3-HYDROXY-2-OXO-4-PHOPHONOXY- BUTYL)-PHOSPHONIC ACID, Glyceraldehyde 3-phosphate dehydrogenase, glycosomal, ... | | Authors: | Castilho, M.S, Pavao, F, Oliva, G. | | Deposit date: | 2003-09-08 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of Trypanosoma cruzi glyceraldehyde-3-phosphate dehydrogenase complexed with an analogue of 1,3-bisphospho-d-glyceric acid.
Eur.J.Biochem., 270, 2003
|
|
8RMJ
 
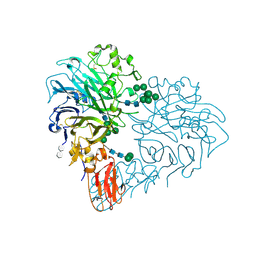 | | Drosophila Semaphorin 2b in complex with glycosaminoglycan mimic SOS | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nourisanami, F, Sobol, M, Rozbesky, D. | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Molecular mechanisms of proteoglycan-mediated semaphorin signaling in axon guidance.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3C6M
 
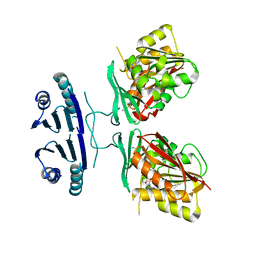 | | Crystal structure of human spermine synthase in complex with spermine and 5-methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, SPERMINE, Spermine synthase | | Authors: | Min, J, Wu, H, Zeng, H, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Pegg, A.E, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-04 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of human spermine synthase: implications of substrate binding and catalytic mechanism.
J.Biol.Chem., 283, 2008
|
|
3C6K
 
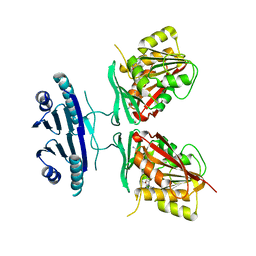 | | Crystal structure of human spermine synthase in complex with spermidine and 5-methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, SPERMIDINE, Spermine synthase | | Authors: | Min, J, Wu, H, Zeng, H, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Pegg, A.E, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-04 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human spermine synthase: implications of substrate binding and catalytic mechanism.
J.Biol.Chem., 283, 2008
|
|
4FL6
 
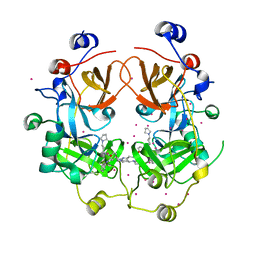 | | Crystal structure of the complex of the 3-MBT repeat domain of L3MBTL3 and UNC1215 | | Descriptor: | Lethal(3)malignant brain tumor-like protein 3, UNKNOWN ATOM OR ION, [2-(phenylamino)benzene-1,4-diyl]bis{[4-(pyrrolidin-1-yl)piperidin-1-yl]methanone} | | Authors: | Zhong, N, Tempel, W, Ravichandran, M, Dong, A, Ingerman, L.A, Graslund, S, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-14 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain.
Nat. Chem. Biol., 9, 2013
|
|
5XQO
 
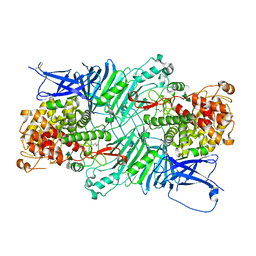 | | Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with tetrameric substrate | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-alpha-L-rhamnopyranose-(1-4)-alpha-D-galactopyranuronic acid-(1-2)-alpha-L-rhamnopyranose, 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-alpha-L-rhamnopyranose-(1-4)-alpha-D-galactopyranuronic acid-(1-2)-alpha-L-rhamnopyranose, CALCIUM ION, ... | | Authors: | Kunishige, Y, Iwai, M, Tada, T, Nishimura, S, Sakamoto, T. | | Deposit date: | 2017-06-07 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of exo-rhamnogalacturonan lyase from Penicillium chrysogenum as a member of polysaccharide lyase family 26
FEBS Lett., 592, 2018
|
|
8RD3
 
 | |
8RDD
 
 | |
5H65
 
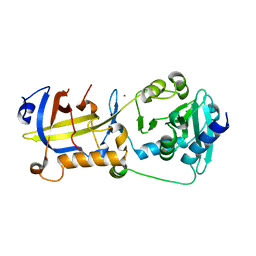 | | Crystal structure of human POT1 and TPP1 | | Descriptor: | Adrenocortical dysplasia protein homolog, Protection of telomeres protein 1, ZINC ION | | Authors: | Chen, C, Wu, J, Lei, M. | | Deposit date: | 2016-11-10 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into POT1-TPP1 interaction and POT1 C-terminal mutations in human cancer.
Nat Commun, 8, 2017
|
|
3CA9
 
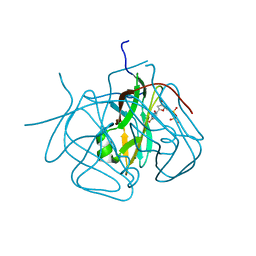 | | Evolution of chlorella virus dUTPase | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine triphosphatase, MAGNESIUM ION | | Authors: | Yamanishi, M, Homma, K, Zhang, Y, Etten, L.V.J, Moriyama, H. | | Deposit date: | 2008-02-19 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallization and crystal-packing studies of Chlorella virus deoxyuridine triphosphatase.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
122D
 
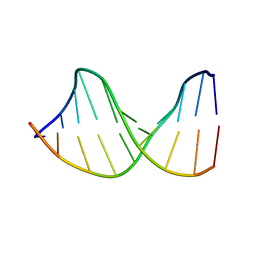 | |
123D
 
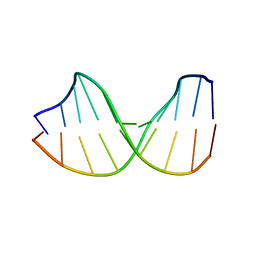 | |
6ZTK
 
 | | Crystal structure of Mialostatin, a gut cystatin from the hard tick Ixodes ricinus | | Descriptor: | FRAGMENT OF TRITON X-100, Mialostatin, SULFATE ION | | Authors: | Busa, M, Rezacova, P, Mares, M. | | Deposit date: | 2020-07-20 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mialostatin, a Novel Midgut Cystatin from Ixodes ricinus Ticks: Crystal Structure and Regulation of Host Blood Digestion.
Int J Mol Sci, 22, 2021
|
|
3CDD
 
 | | Crystal structure of prophage MuSo2, 43 kDa tail protein from Shewanella oneidensis | | Descriptor: | Prophage MuSo2, 43 kDa tail protein | | Authors: | Chang, C, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-26 | | Release date: | 2008-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of prophage MuSo2, 43 kDa tail protein from Shewanella oneidensis.
To be Published
|
|
5XQG
 
 | | Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-alpha-L-rhamnopyranose, CALCIUM ION, Pcrglx protein | | Authors: | Kunishige, Y, Iwai, M, Tada, T, Nishimura, S, Sakamoto, T. | | Deposit date: | 2017-06-07 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal structure of exo-rhamnogalacturonan lyase from Penicillium chrysogenum as a member of polysaccharide lyase family 26
FEBS Lett., 592, 2018
|
|
7ABT
 
 | |
4ORU
 
 | | Three-dimensional structure of the C65A mutant of Human lipocalin-type Prostaglandin D Synthase holo-form second space group | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, Prostaglandin-H2 D-isomerase | | Authors: | Perduca, M, Bovi, M, Bertinelli, M, Bertini, E, Destefanis, L, Carrizo, M.E, Capaldi, S, Monaco, H.L. | | Deposit date: | 2014-02-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High-resolution structures of mutants of residues that affect access to the ligand-binding cavity of human lipocalin-type prostaglandin D synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
