3WL4
 
 | | N,N'-diacetylchitobiose deacetylase (Se-derivative) from Pyrococcus furiosus | | Descriptor: | CADMIUM ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Uegaki, K. | | Deposit date: | 2013-11-07 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Expression from engineered Escherichia coli chromosome and crystallographic study of archaeal N,N'-diacetylchitobiose deacetylase
Febs J., 281, 2014
|
|
3QZX
 
 | | 3D Structure of ferric methanosarcina acetivorans protoglobin Y61A mutant with unknown ligand | | Descriptor: | GLYCEROL, Methanosarcina acetivorans protoglobin, PHOSPHATE ION, ... | | Authors: | Pesce, A, Tilleman, L, Dewilde, S, Ascenzi, P, Coletta, M, Ciaccio, C, Bruno, S, Moens, L, Bolognesi, M, Nardini, M. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural heterogeneity and ligand gating in ferric methanosarcina acetivorans protoglobin mutants.
Iubmb Life, 63, 2011
|
|
3WKG
 
 | | Crystal structure of cellobiose 2-epimerase in complex with glucosylmannose | | Descriptor: | CHLORIDE ION, Cellobiose 2-epimerase, PHOSPHATE ION, ... | | Authors: | Fujiwara, T, Saburi, W, Tanaka, I, Yao, M. | | Deposit date: | 2013-10-21 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Insights into the Epimerization of beta-1,4-Linked Oligosaccharides Catalyzed by Cellobiose 2-Epimerase, the Sole Enzyme Epimerizing Non-anomeric Hydroxyl Groups of Unmodified Sugars
J.Biol.Chem., 289, 2014
|
|
3WMP
 
 | | Crystal structure of SLL-2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kita, A, Jimbo, M, Sakai, R, Morimoto, Y, Miki, K. | | Deposit date: | 2013-11-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a symbiosis-related lectin from octocoral.
Glycobiology, 25, 2015
|
|
3WN1
 
 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase in complex with xylotriose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
3WP3
 
 | |
3WVO
 
 | | Crystal structure of Thermobifida fusca Cse1 | | Descriptor: | CRISPR-associated protein, Cse1 family | | Authors: | Yuan, Y.A, Tay, M. | | Deposit date: | 2014-06-02 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structure of Thermobifida fusca Cse1 reveals target DNA binding site.
Protein Sci., 24, 2015
|
|
3WWP
 
 | | S-selective hydroxynitrile lyase from Baliospermum montanum (apo2) | | Descriptor: | (S)-hydroxynitrile lyase, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Nakano, S, Dadashipour, M, Asano, Y. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of hydroxynitrile lyase from Baliospermum montanum with crystal structure, molecular dynamics and enzyme kinetics
Biochim.Biophys.Acta, 1844, 2014
|
|
3WXP
 
 | |
3WAP
 
 | | Crystal structure of Atg13 LIR-fused human LC3C_8-125 | | Descriptor: | Autophagy-related protein 13, Microtubule-associated proteins 1A/1B light chain 3C | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WAM
 
 | | Crystal structure of human LC3C_8-125 | | Descriptor: | CITRIC ACID, Microtubule-associated proteins 1A/1B light chain 3C | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
2OAA
 
 | | Restriction endonuclease MvaI-cognate DNA substrate complex | | Descriptor: | 5'-D(*CP*AP*TP*CP*CP*AP*GP*GP*TP*AP*C)-3', 5'-D(*GP*GP*TP*AP*CP*CP*TP*GP*GP*AP*T)-3', CALCIUM ION, ... | | Authors: | Kaus-Drobek, M, Czapinska, H, Sokolowska, M, Tamulaitis, G, Szczepanowski, R.H, Urbanke, K, Siksnys, V, Bochtler, M. | | Deposit date: | 2006-12-15 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Restriction endonuclease MvaI is a monomer that recognizes its target sequence asymmetrically.
Nucleic Acids Res., 35, 2007
|
|
3K4S
 
 | | The structure of the catalytic domain of human PDE4d with 4-(3-Butoxy-4-methoxyphenyl)methyl-2-imidazolidone | | Descriptor: | (4R)-4-(3-butoxy-4-methoxybenzyl)imidazolidin-2-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Crawley, L, Cheng, R.K.Y, Wood, M, Barker, J, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-10-06 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of the catalytic domain of human PDE4d with 4-(3-Butoxy-4-methoxyphenyl)methyl-2-imidazolidone
To be Published
|
|
3K6L
 
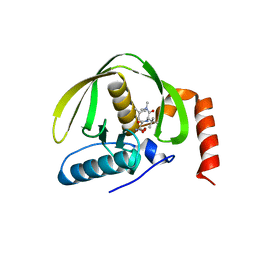 | | The structure of E.coli peptide deformylase (PDF) in complex with peptidomimetic ligand BB2827 | | Descriptor: | (2S,3R)-N~4~-[(1S)-1-(dimethylcarbamoyl)-2,2-dimethylpropyl]-N~1~,2-dihydroxy-3-(2-methylpropyl)butanediamide, NICKEL (II) ION, Peptide deformylase | | Authors: | Cheng, R.K.Y, Crawley, L, Wood, M, Barker, J, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-10-09 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of E.coli peptide deformylase (PDF) in complex with peptidomimetic ligand BB2827
To be Published
|
|
4YJT
 
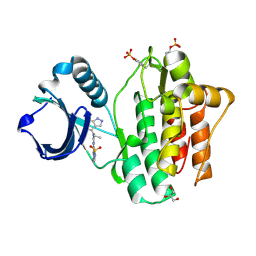 | | THE KINASE DOMAIN OF HUMAN SPLEEN TYROSINE (SYK) IN COMPLEX WITH GTC000233 | | Descriptor: | GLYCEROL, N~2~-(1,1-dioxido-2,3-dihydro-1,2-benzothiazol-6-yl)-N~4~-ethyl-5-fluoro-N~4~-(1H-indazol-4-yl)pyrimidine-2,4-diamine, Tyrosine-protein kinase SYK | | Authors: | Somers, D.O, Neu, M, Stuckey, J. | | Deposit date: | 2015-03-03 | | Release date: | 2015-09-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of GSK143, a highly potent, selective and orally efficacious spleen tyrosine kinase inhibitor.
Bioorg. Med. Chem. Lett., 21, 2011
|
|
3K7X
 
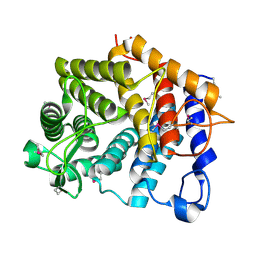 | | Crystal structure of the Lin0763 protein from Listeria innocua. Northeast Structural Genomics Consortium Target LkR23. | | Descriptor: | Lin0763 protein, PHOSPHATE ION | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Maglaqui, M, Ciccosanti, C, Buchwald, W.A, Sahdev, S, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-13 | | Release date: | 2009-10-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Crystal structure of the Lin0763 protein from Listeria innocua.
To be Published
|
|
3ZJG
 
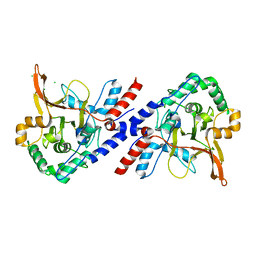 | | A20 OTU domain with irreversibly oxidised Cys103 from 60 min H2O2 soak. | | Descriptor: | CHLORIDE ION, TUMOR NECROSIS FACTOR ALPHA-INDUCED PROTEIN 3 | | Authors: | Kulathu, Y, Garcia, F.J, Mevissen, T.E.T, Busch, M, Arnaudo, N, Carroll, K.S, Barford, D, Komander, D. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Regulation of A20 and Other Otu Deubiquitinases by Reversible Oxidation
Nat.Commun., 4, 2013
|
|
1FWX
 
 | | CRYSTAL STRUCTURE OF NITROUS OXIDE REDUCTASE FROM P. DENITRIFICANS | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Brown, K, Djinovic-Carugo, K, Haltia, T, Cabrito, I, Saraste, M, Moura, J.J, Moura, I, Tegoni, M, Cambillau, C. | | Deposit date: | 2000-09-25 | | Release date: | 2001-09-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Revisiting the Catalytic CuZ Cluster of Nitrous Oxide (N2O) Reductase. Evidence of a Bridging Inorganic Sulfur
J.Biol.Chem., 275, 2000
|
|
3ZVE
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 84 | | Descriptor: | 3C PROTEASE, O-tert-butyl-N-[(9H-fluoren-9-ylmethoxy)carbonyl]-L-threonyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3JQQ
 
 | |
3ZWX
 
 | | Crystal structure of ADP-ribosyl cyclase complexed with 8-bromo-ADP- ribose | | Descriptor: | ADP-RIBOSYL CYCLASE, CHLORIDE ION, [(2R,3S,4R,5R)-5-(6-amino-8-bromo-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3S,4S)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
To be Published
|
|
3ZY5
 
 | | Crystal structure of POFUT1 in complex with GDP-fucose (crystal-form-I) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GUANOSINE-5'-DIPHOSPHATE-BETA-L-FUCOPYRANOSE, PUTATIVE GDP-FUCOSE PROTEIN O-FUCOSYLTRANSFERASE 1, ... | | Authors: | Lira-Navarrete, E, Valero-Gonzalez, J, Villanueva, R, Martinez-Julvez, M, Tejero, T, Merino, P, Panjikar, S, Hurtado-Guerrero, R. | | Deposit date: | 2011-08-17 | | Release date: | 2011-09-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Insights Into the Mechanism of Protein O-Fucosylation.
Plos One, 6, 2011
|
|
3JTK
 
 | | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD90055 | | Descriptor: | (2R)-3-benzyl-2-(2-bromo-4-hydroxy-5-methoxyphenyl)-1,3-thiazolidin-4-one, Glycylpeptide N-tetradecanoyltransferase 1, TETRADECANOYL-COA | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Lin, Y.-H, Kania, A, Ravichandran, M, Kozieradzki, I, Cossar, D, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Wyatt, P.G, Ferguson, M.A.J, Frearson, J.A, Brand, S.Y, Robinson, D.A, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-12 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD90055
To be Published
|
|
3ZG3
 
 | | STEROL 14-ALPHA DEMETHYLASE (CYP51)FROM TRYPANOSOMA CRUZI IN COMPLEX WITH THE PYRIDINE INHIBITOR N-(1-(5-(trifluoromethyl)(pyridin-2-yl)) piperidin-4yl)-N-(4-(trifluoromethyl)phenyl)pyridin-3-amine (EPL- BS967, UDD) | | Descriptor: | N-[4-(trifluoromethyl)phenyl]-N-[1-[5-(trifluoromethyl)pyridin-2-yl]piperidin-4-yl]pyridin-3-amine, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Hargrove, T.Y, Wawrzak, Z, Keenan, M, Chatelain, E, Lepesheva, G.I. | | Deposit date: | 2012-12-14 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Complexes of Trypanosoma Cruzi Sterol 14Alpha-Demethylase (Cyp51) with Two Pyridine-Based Drug Candidates for Chagas Disease: Structural Basis for Pathogen-Selectivity
J.Biol.Chem., 288, 2013
|
|
3ZEZ
 
 | | Phage dUTPases control transfer of virulence genes by a proto- oncogenic G protein-like mechanism.(Staphylococcus bacteriophage 80alpha dUTPase with dUPNHPP). | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPASE, MAGNESIUM ION, ... | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
