4KF9
 
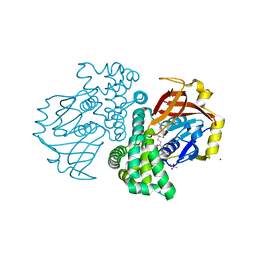 | | Crystal structure of a glutathione transferase family member from ralstonia solanacearum, target efi-501780, with bound gsh coordinated to a zinc ion, ordered active site | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione s-transferase protein, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-26 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a glutathione transferase family member from ralstonia solanacearum, target efi-501780, with bound gsh coordinated to a zinc ion, ordered active site
To be Published
|
|
2AHM
 
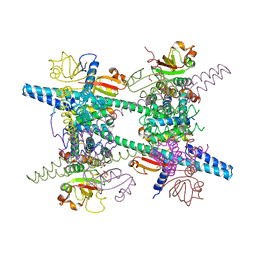 | | Crystal structure of SARS-CoV super complex of non-structural proteins: the hexadecamer | | Descriptor: | GLYCEROL, Replicase polyprotein 1ab, heavy chain, ... | | Authors: | Zhai, Y.J, Sun, F, Bartlam, M, Rao, Z. | | Deposit date: | 2005-07-28 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer
NAT.STRUCT.MOL.BIOL., 12, 2005
|
|
2AP3
 
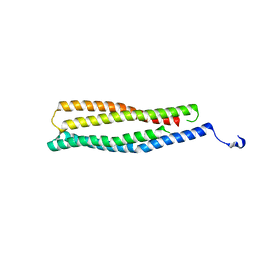 | | 1.6 A Crystal Structure of a Conserved Protein of Unknown Function from Staphylococcus aureus | | Descriptor: | conserved hypothetical protein | | Authors: | Zhang, R, Zhou, M, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-15 | | Release date: | 2005-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.6A crystal structure of a conserved hypothetical protein from Staphylococcus aureus MW2
To be Published
|
|
1ZF0
 
 | | B-DNA | | Descriptor: | 5'-D(*CP*CP*GP*TP*TP*AP*AP*CP*GP*G)-3', CALCIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4KNP
 
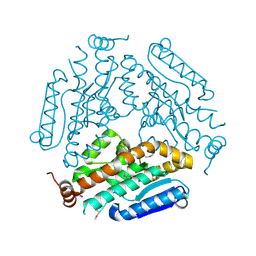 | | Crystal Structure Of a Putative enoyl-coA hydratase (PSI-NYSGRC-019597) from Mycobacterium avium paratuberculosis K-10 | | Descriptor: | enoyl-CoA hydratase | | Authors: | Kumar, P.R, Ahmed, M, Attonito, J, Bhosle, R, Chamala, S, Chowdhury, S, Glenn, A.S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Wasserman, S.R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-05-10 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal structure of a Putative enoyl-coA hydratase from Mycobacterium avium paratuberculosis K-10
to be published
|
|
1YV2
 
 | | Hepatitis C virus NS5B RNA-dependent RNA Polymerase genotype 2a | | Descriptor: | GLYCEROL, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the RNA-dependent RNA Polymerase Genotype 2a of Hepatitis C Virus Reveal Two Conformations and Suggest Mechanisms of Inhibition by Non-nucleoside Inhibitors
J.Biol.Chem., 280, 2005
|
|
4K33
 
 | | Crystal Structure of FGF Receptor 3 (FGFR3) Kinase Domain Harboring the K650E Mutation, a Gain-of-Function Mutation Responsible for Thanatophoric Dysplasia Type II and Spermatocytic Seminoma | | Descriptor: | Fibroblast growth factor receptor 3, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Huang, Z, Chen, H, Mohammadi, M. | | Deposit date: | 2013-04-10 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3405 Å) | | Cite: | Structural Mimicry of A-Loop Tyrosine Phosphorylation by a Pathogenic FGF Receptor 3 Mutation.
Structure, 21, 2013
|
|
3PY4
 
 | | Crystal structure of bovine lactoperoxidase in complex with paracetamol at 2.4A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pandey, N, Sing, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-12-11 | | Release date: | 2011-01-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of bovine lactoperoxidase in complex with paracetamol at 2.4A resolution
To be published
|
|
1YVZ
 
 | | Hepatitis C Virus RNA Polymerase Genotype 2a In Complex With Non- Nucleoside Analogue Inhibitor | | Descriptor: | 3-[(2,4-DICHLOROBENZOYL)(ISOPROPYL)AMINO]-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
3PYB
 
 | | Crystal structure of ent-copalyl diphosphate synthase from Arabidopsis thaliana in complex with 13-aza-13,14-dihydrocopalyl diphosphate | | Descriptor: | 2-(methyl{2-[(1S,4aS,8aS)-5,5,8a-trimethyl-2-methylidenedecahydronaphthalen-1-yl]ethyl}amino)ethyl trihydrogen diphosphate, Ent-copalyl diphosphate synthase, chloroplastic, ... | | Authors: | Koksal, M, Christianson, D.W. | | Deposit date: | 2010-12-12 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.759 Å) | | Cite: | Structure of ent-Copalyl Diphosphate Synthase from Arabidopsis thaliana, a Protonation-Dependent Diterpene Cyclase
To be Published
|
|
4KPV
 
 | | Crystal structure of the complex of ribosome inactivating protein from Momordica balsamina with Pyrimidine-2,4(1H,3H)-dione at 2.57 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, URACIL, rRNA N-glycosidase | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-05-14 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structure of the complex of ribosome inactivating protein from Momordica balsamina with Pyrimidine-2,4(1H,3H)-dione at 2.57 A resolution
To be Published
|
|
1ZLR
 
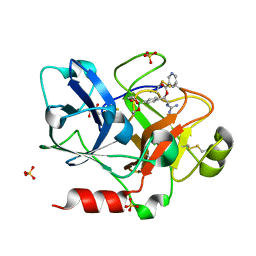 | | Factor XI catalytic domain complexed with 2-guanidino-1-(4-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)ethyl nicotinate | | Descriptor: | (1R)-2-{[AMINO(IMINO)METHYL]AMINO}-1-{4-[(4R)-4-(HYDROXYMETHYL)-1,3,2-DIOXABOROLAN-2-YL]PHENYL}ETHYL NICOTINATE, Coagulation factor XI, GLYCEROL, ... | | Authors: | Lazarova, T.I, Jin, L, Rynkiewicz, M, Gorga, J.C, Bibbins, F, Meyers, H.V, Babine, R, Strickler, J. | | Deposit date: | 2005-05-09 | | Release date: | 2006-05-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synthesis and in vitro biological evaluation of aryl boronic acids as potential inhibitors of factor XIa.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3Q28
 
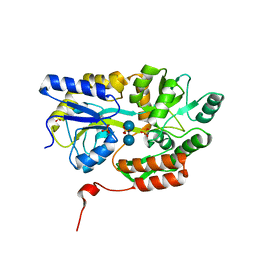 | | Cyrstal structure of human alpha-synuclein (58-79) fused to maltose binding protein (MBP) | | Descriptor: | Maltose-binding periplasmic protein/alpha-synuclein chimeric protein, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhao, M, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2010-12-19 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of segments of alpha-synuclein fused to maltose-binding protein suggest intermediate states during amyloid formation
Protein Sci., 20, 2011
|
|
3Q0I
 
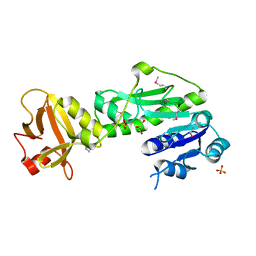 | | Methionyl-tRNA formyltransferase from Vibrio cholerae | | Descriptor: | GLYCEROL, Methionyl-tRNA formyltransferase, SULFATE ION | | Authors: | Osipiuk, J, Gu, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-15 | | Release date: | 2010-12-29 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Methionyl-tRNA formyltransferase from Vibrio cholerae.
To be Published
|
|
1ZMC
 
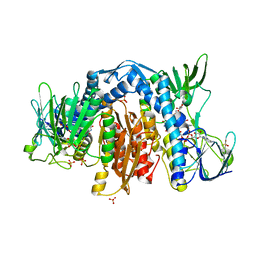 | | Crystal Structure of Human dihydrolipoamide dehydrogenase complexed to NAD+ | | Descriptor: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Brautigam, C.A, Chuang, J.L, Tomchick, D.R, Machius, M, Chuang, D.T. | | Deposit date: | 2005-05-10 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal Structure of Human Dihydrolipoamide Dehydrogenase: NAD(+)/NADH Binding and the Structural Basis of Disease-causing Mutations
J.Mol.Biol., 350, 2005
|
|
1YUY
 
 | | HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE GENOTYPE 2a | | Descriptor: | RNA-Dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
3Q3W
 
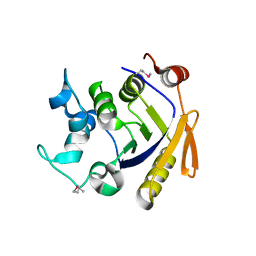 | | Isopropylmalate isomerase small subunit from Campylobacter jejuni. | | Descriptor: | 3-isopropylmalate dehydratase small subunit | | Authors: | Osipiuk, J, Gu, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-22 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Isopropylmalate isomerase small subunit from Campylobacter jejuni.
To be Published
|
|
3PHW
 
 | | OTU Domain of Crimean Congo Hemorrhagic Fever Virus in complex with Ubiquitin | | Descriptor: | ETHANAMINE, RNA-directed RNA polymerase L, Ubiquitin-40S ribosomal protein S27a | | Authors: | Akutsu, M, Ye, Y, Virdee, S, Komander, D. | | Deposit date: | 2010-11-04 | | Release date: | 2011-02-02 | | Last modified: | 2014-08-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4K23
 
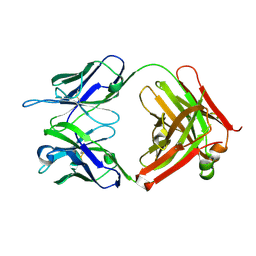 | | Structure of anti-uPAR Fab ATN-658 | | Descriptor: | anti-uPAR antibody, heavy chain, light chain | | Authors: | Yuan, C, Huang, M, Chen, L. | | Deposit date: | 2013-04-08 | | Release date: | 2014-02-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a New Epitope in uPAR as a Target for the Cancer Therapeutic Monoclonal Antibody ATN-658, a Structural Homolog of the uPAR Binding Integrin CD11b ( alpha M)
Plos One, 9, 2014
|
|
1Z3M
 
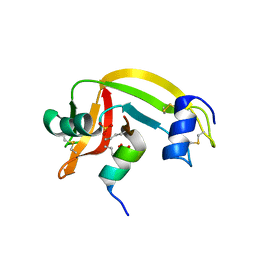 | | Crystal structure of mutant Ribonuclease S (F8Nva) | | Descriptor: | Ribonuclease pancreatic, S-Peptide, S-protein, ... | | Authors: | Das, M, Vasudeva Rao, B, Ghosh, S, Varadarajan, R. | | Deposit date: | 2005-03-14 | | Release date: | 2005-03-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Attempts to delineate the relative contributions of changes in hydrophobicity and packing to changes in stability of ribonuclease S mutants.
Biochemistry, 44, 2005
|
|
1Z4H
 
 | | The response regulator TorI belongs to a new family of atypical excisionase | | Descriptor: | Tor inhibition protein | | Authors: | Elantak, L, Ansaldi, M, Guerlesquin, F, Mejean, V, Morelli, X. | | Deposit date: | 2005-03-16 | | Release date: | 2005-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and genetic analyses reveal a key role in prophage excision for the TorI response regulator inhibitor
J.Biol.Chem., 280, 2005
|
|
3PM7
 
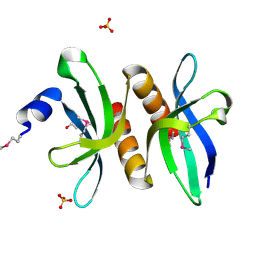 | | Crystal Structure of EF_3132 protein from Enterococcus faecalis at the resolution 2A, Northeast Structural Genomics Consortium Target EfR184 | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, Uncharacterized protein | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-11-16 | | Release date: | 2010-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Northeast Structural Genomics Consortium Target EfR184
To be Published
|
|
3PMX
 
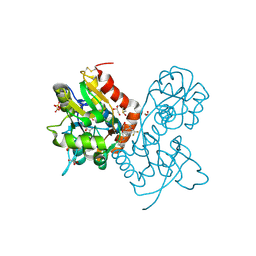 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | Authors: | Maclean, J.K.F, Jamieson, C, Brown, C.I, Campbell, R.A, Gillen, K.J, Gillespie, J, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure based evolution of a novel series of positive modulators of the AMPA receptor.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4KES
 
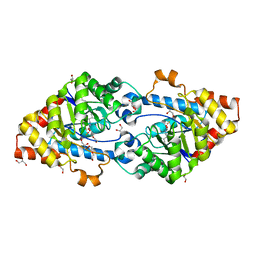 | | Crystal structure of SsoPox W263T | | Descriptor: | 1,2-ETHANEDIOL, Aryldialkylphosphatase, COBALT (II) ION, ... | | Authors: | Gotthard, G, Hiblot, J, Chabriere, E, Elias, M. | | Deposit date: | 2013-04-26 | | Release date: | 2013-10-02 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Differential Active Site Loop Conformations Mediate Promiscuous Activities in the Lactonase SsoPox.
Plos One, 8, 2013
|
|
1YZW
 
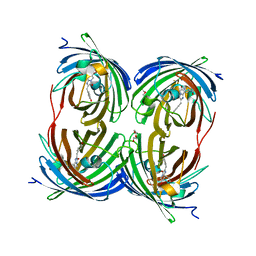 | | The 2.1A Crystal Structure of the Far-red Fluorescent Protein HcRed: Inherent Conformational Flexibility of the Chromophore | | Descriptor: | DI(HYDROXYETHYL)ETHER, GFP-like non-fluorescent chromoprotein | | Authors: | Wilmann, P.G, Petersen, J, Pettikiriarachchi, A, Buckle, A.M, Devenish, R.J, Prescott, M, Rossjohn, J. | | Deposit date: | 2005-02-28 | | Release date: | 2005-05-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1A Crystal Structure of the Far-red Fluorescent Protein HcRed: Inherent Conformational Flexibility of the Chromophore
J.Mol.Biol., 349, 2005
|
|
