6TPS
 
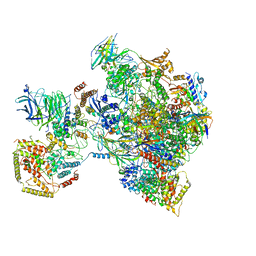 | | early intermediate RNA Polymerase I Pre-initiation complex - eiPIC | | Descriptor: | DNA foreign, DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, ... | | Authors: | Pilsl, M, Engel, C. | | Deposit date: | 2019-12-14 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of RNA polymerase I pre-initiation complex formation and promoter melting.
Nat Commun, 11, 2020
|
|
6TRH
 
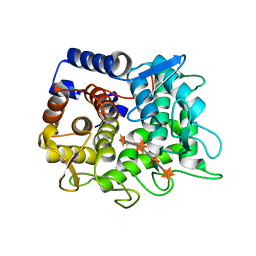 | |
3ZFM
 
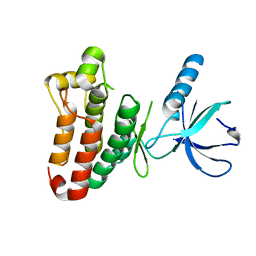 | | Crystal structure of EphB2 | | Descriptor: | EPHRIN TYPE-B RECEPTOR 2 | | Authors: | Debreczeni, J.E, Overman, R, Truman, C, McAlister, M, Attwood, T.K. | | Deposit date: | 2012-12-12 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Completing the Structural Family Portrait of the Human Ephb Tyrosine Kinase Domains
Protein Sci., 23, 2014
|
|
6TT0
 
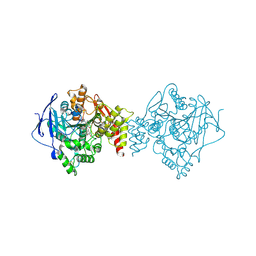 | | Crystal structure of a potent and reversible dual binding site Acetylcholinesterase chiral inhibitor | | Descriptor: | (1~{R},3~{S})-~{N}-(6,7-dimethoxy-2-oxidanylidene-chromen-3-yl)-3-[(phenylmethyl)amino]cyclohexane-1-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | de la Mora, E, Mangiatordi, G.F, Belviso, B.D, Caliandro, R, Colletier, J.P, Catto, M. | | Deposit date: | 2019-12-22 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.80003023 Å) | | Cite: | Chiral Separation, X-ray Structure, and Biological Evaluation of a Potent and Reversible Dual Binding Site AChE Inhibitor.
Acs Med.Chem.Lett., 11, 2020
|
|
3ZEW
 
 | | Crystal structure of EphB4 in complex with staurosporine | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, STAUROSPORINE, SULFATE ION | | Authors: | Debreczeni, J.E, Overman, R, Truman, C, McAlister, M, Attwood, T.K. | | Deposit date: | 2012-12-07 | | Release date: | 2013-12-25 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Completing the Structural Family Portrait of the Human Ephb Tyrosine Kinase Domains
Protein Sci., 23, 2014
|
|
6TT8
 
 | | Haddock model of NDM-1/morin complex | | Descriptor: | 2-[2,4-bis(oxidanyl)phenyl]-3,5,7-tris(oxidanyl)chromen-4-one, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | Deposit date: | 2019-12-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
6TQX
 
 | |
6TQV
 
 | |
7JOZ
 
 | | Crystal structure of dopamine D1 receptor in complex with G protein and a non-catechol agonist | | Descriptor: | 6-{4-[(furo[3,2-c]pyridin-4-yl)oxy]-2-methylphenyl}-1,5-dimethylpyrimidine-2,4(1H,3H)-dione, Endolysin,D(1A) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Sun, B, Feng, D, Chu, M.L, Fish, I, Kelm, S, Lebon, F, Lovera, S, Valade, A, Wood, M, Ceska, T, Kobilka, T.S, Sands, Z, Kobilka, B.K. | | Deposit date: | 2020-08-07 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of dopamine D1 receptor in complex with G protein and a non-catechol agonist.
Nat Commun, 12, 2021
|
|
6TSH
 
 | | Beta-galactosidase in complex with deoxygalacto-nojirimycin | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, Beta-galactosidase, MAGNESIUM ION | | Authors: | Saur, M, Hartshorn, M.J, Dong, J, Reeks, J, Bunkoczi, G, Jhoti, H, Williams, P.A. | | Deposit date: | 2019-12-20 | | Release date: | 2020-01-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Fragment-based drug discovery using cryo-EM.
Drug Discov Today, 25, 2020
|
|
6TTH
 
 | | PKM2 in complex with L-threonine | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Pyruvate kinase PKM, THREONINE | | Authors: | Saur, M, Hartshorn, M.J, Dong, J, Reeks, J, Bunkoczi, G, Jhoti, H, Williams, P.A. | | Deposit date: | 2019-12-27 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Fragment-based drug discovery using cryo-EM.
Drug Discov Today, 25, 2020
|
|
6TTQ
 
 | | PKM2 in complex with Compound 10 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 1-propan-2-yl-3-pyridin-4-yl-urea, Pyruvate kinase PKM | | Authors: | Saur, M, Hartshorn, M.J, Dong, J, Reeks, J, Bunkoczi, G, Jhoti, H, Williams, P.A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Fragment-based drug discovery using cryo-EM.
Drug Discov Today, 25, 2020
|
|
7EG2
 
 | | Crystal structure of the apoAequorin complex with (S)-daCTZ | | Descriptor: | (2~{S})-2-(hydroxymethyl)-6-(4-hydroxyphenyl)-2-[(4-hydroxyphenyl)methyl]-4-(phenylmethyl)-3~{H}-inden-1-one, Aequorin-2 | | Authors: | Tomabechi, Y, Shirouzu, M. | | Deposit date: | 2021-03-24 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Chiral deaza-coelenterazine analogs for probing a substrate-binding site in the Ca2+-binding photoprotein aequorin.
Plos One, 16, 2021
|
|
7EG3
 
 | | Crystal structure of the apoAequorin complex with (S)-HM-daCTZ | | Descriptor: | (2~{S})-6-(4-hydroxyphenyl)-2-[(4-hydroxyphenyl)methyl]-4-(phenylmethyl)-2,3-dihydroinden-1-one, Aequorin-2 | | Authors: | Tomabechi, Y, Shirouzu, M. | | Deposit date: | 2021-03-24 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Chiral deaza-coelenterazine analogs for probing a substrate-binding site in the Ca2+-binding photoprotein aequorin.
Plos One, 16, 2021
|
|
6TTZ
 
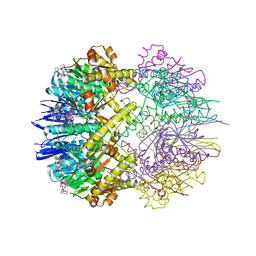 | | Structure of the ClpP:ADEP4-complex from Staphylococcus aureus (open state) | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, N-[(2S)-3-(3,5-difluorophenyl)-1-[[(3S,9S,13S,15R,19S,22S)-15,19-dimethyl-2,8,12,18,21-pentaoxo-11-oxa-1,7,17,20-tetrazatetracyclo[20.4.0.03,7.013,17]hexacosan-9-yl]amino]-1-oxopropan-2-yl]heptanamide | | Authors: | Malik, I.T, Pereira, R, Vielberg, M.-T, Mayer, C, Straetener, J, Thomy, D, Famulla, K, Castro, H.C, Sass, P, Groll, M, Broetz-Oesterheldt, H. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional Characterisation of ClpP Mutations Conferring Resistance to Acyldepsipeptide Antibiotics in Firmicutes.
Chembiochem, 21, 2020
|
|
6TUG
 
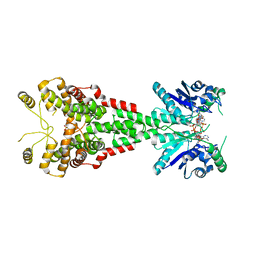 | |
7E0H
 
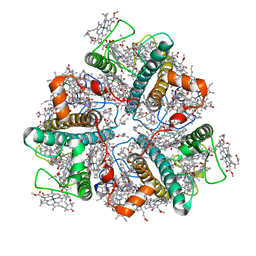 | | LHCII-1 in the state transition supercomplex PSI-LHCI-LHCII from the LhcbM1 lacking mutant of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Li, A.J, Liu, Z.F, Li, M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-06-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural basis of LhcbM5-mediated state transitions in green algae.
Nat.Plants, 7, 2021
|
|
7DZ8
 
 | | State transition supercomplex PSI-LHCI-LHCII from the LhcbM1 lacking mutant of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Li, A.J, Liu, Z.F, Li, M. | | Deposit date: | 2021-01-23 | | Release date: | 2021-06-30 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis of LhcbM5-mediated state transitions in green algae.
Nat.Plants, 7, 2021
|
|
7E0J
 
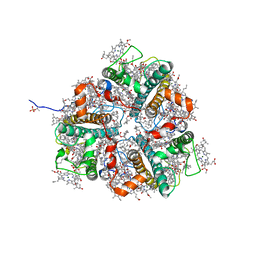 | | LHCII-1 in the state transition supercomplex PSI-LHCI-LHCII from the double phosphatase mutant pph1;pbcp of Chlamydomonas reinhardti. | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Li, A.J, Liu, Z.F, Li, M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-06-30 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis of LhcbM5-mediated state transitions in green algae.
Nat.Plants, 7, 2021
|
|
7EGV
 
 | | Acetolactate Synthase from Trichoderma harzianum with inhibitor harzianic acid | | Descriptor: | (2S)-3-methyl-2-[[(2S,4R)-1-methyl-4-[(2E,4E)-octa-2,4-dienoyl]-3,5-bis(oxidanylidene)pyrrolidin-2-yl]methyl]-2-oxidanyl-butanoic acid, 1,2-ETHANEDIOL, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ... | | Authors: | Zang, X, Xie, L, Chen, M, Tang, Y, Zhou, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Harzianic Acid from Trichoderma afroharzianum Is a Natural Product Inhibitor of Acetohydroxyacid Synthase.
J.Am.Chem.Soc., 2021
|
|
7E2E
 
 | | Crystal structure of the Estrogen-Related Receptor alpha (ERRalpha) ligand-binding domain (LBD) in complex with an agonist DS45500853 and a PGC-1alpha peptide | | Descriptor: | 1-[4-(3-tert-butyl-4-oxidanyl-phenoxy)phenyl]ethanone, IODIDE ION, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, ... | | Authors: | Ito, S, Shinozuka, T, Kimura, T, Izumi, M, Wakabayashi, K. | | Deposit date: | 2021-02-05 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of a Novel Class of ERR alpha Agonists.
Acs Med.Chem.Lett., 12, 2021
|
|
7DZ7
 
 | | State transition supercomplex PSI-LHCI-LHCII from double phosphatase mutant pph1;pbcp of green alga Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Li, A.J, Liu, Z.F, Li, M. | | Deposit date: | 2021-01-23 | | Release date: | 2021-06-30 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis of LhcbM5-mediated state transitions in green algae.
Nat.Plants, 7, 2021
|
|
7JQQ
 
 | | The bacteriophage Phi-29 viral genome packaging motor assembly | | Descriptor: | DNA (60-MER), DNA packaging protein, MAGNESIUM ION, ... | | Authors: | White, M.A, Woodson, M, Morais, M.C. | | Deposit date: | 2020-08-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | A viral genome packaging motor transitions between cyclic and helical symmetry to translocate dsDNA.
Sci Adv, 7, 2021
|
|
6SYE
 
 | |
7E0I
 
 | | LHCII-2 in the state transition supercomplex PSI-LHCI-LHCII from the LhcbM1 lacking mutant of Chlamydomonas reinhardti | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Li, A.J, Liu, Z.F, Li, M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-06-30 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural basis of LhcbM5-mediated state transitions in green algae.
Nat.Plants, 7, 2021
|
|
