1BEG
 
 | | STRUCTURE OF FUNGAL ELICITOR, NMR, 18 STRUCTURES | | Descriptor: | BETA-ELICITIN CRYPTOGEIN | | Authors: | Fefeu, S, Bouaziz, S, Huet, J.-C, Pernollet, J.-C, Guittet, E. | | Deposit date: | 1996-11-26 | | Release date: | 1997-12-03 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of beta cryptogein, a beta elicitin secreted by a phytopathogenic fungus Phytophthora cryptogea.
Protein Sci., 6, 1997
|
|
6ZOP
 
 | | Structure of the cysteine-rich domain of PiggyMac, a domesticated PiggyBac transposase involved in programmed genome rearrangements | | Descriptor: | DDE_Tnp_1_7 domain-containing protein, ZINC ION | | Authors: | Bessa, L, Guerineau, M, Moriau, S, Lescop, E, Bontems, F, Mathy, N, Guittet, E, Bischerour, J, Betermier, M, Morellet, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | SOLUTION NMR | | Cite: | The unusual structure of the PiggyMac cysteine-rich domain reveals zinc finger diversity in PiggyBac-related transposases.
Mob DNA, 12, 2021
|
|
5LME
 
 | | Specific-DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase | | Descriptor: | ZINC ION, piggyBac transposase | | Authors: | Morellet, N, Taylor, J.A, Wieninger, S, Moriau, S, Li, X, Lescop, E, Mathy, N, Bischerour, J, Betermier, M, Bardiaux, B, Nilges, M, Craig, N.L, Hickman, A.B, Dyda, F, Guittet, E. | | Deposit date: | 2016-07-30 | | Release date: | 2017-12-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase.
Nucleic Acids Res., 46, 2018
|
|
3ALC
 
 | | ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR DNA-BINDING DOMAIN FROM ASPERGILLUS NIDULANS | | Descriptor: | PROTEIN (ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR), ZINC ION | | Authors: | Cerdan, R, Cahuzac, B, Felenbok, B, Guittet, E. | | Deposit date: | 1999-03-11 | | Release date: | 2000-05-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of AlcR (1-60) provides insight in the unusual DNA binding properties of this zinc binuclear cluster protein.
J.Mol.Biol., 295, 2000
|
|
6TTA
 
 | | Haddock model of NDM-1/quercetin complex | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | Deposit date: | 2019-12-26 | | Release date: | 2021-01-13 | | Last modified: | 2021-03-17 | | Method: | SOLUTION NMR | | Cite: | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
6TT8
 
 | | Haddock model of NDM-1/morin complex | | Descriptor: | 2-[2,4-bis(oxidanyl)phenyl]-3,5,7-tris(oxidanyl)chromen-4-one, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | Deposit date: | 2019-12-25 | | Release date: | 2021-01-13 | | Last modified: | 2021-03-17 | | Method: | SOLUTION NMR | | Cite: | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
6TTC
 
 | | Haddock model of NDM-1/myricetin complex | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | Deposit date: | 2019-12-26 | | Release date: | 2021-01-13 | | Last modified: | 2021-03-17 | | Method: | SOLUTION NMR | | Cite: | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
2LS1
 
 | | Structure of Sviceucin, an antibacterial type I lasso peptide from Streptomyces sviceus | | Descriptor: | Uncharacterized protein | | Authors: | Li, Y, Ducasse, R, Blond, A, Zirah, S, Goulard, C, Lescop, E, Guittet, E, Pernodet, J, Rebuffat, S. | | Deposit date: | 2012-04-18 | | Release date: | 2012-08-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and biosynthesis of Sviceucin, an antibacterial type I lasso peptide from Streptomyces sviceus
To be Published
|
|
1D2D
 
 | | Hamster EprS second repeated element. NMR, 5 structures | | Descriptor: | TRNA SYNTHETASE | | Authors: | Cahuzac, B, Berthonneau, E, Birlirakis, N, Guittet, E, Mirande, M. | | Deposit date: | 1999-09-23 | | Release date: | 2000-05-24 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | A recurrent RNA-binding domain is appended to eukaryotic aminoacyl-tRNA synthetases.
EMBO J., 19, 2000
|
|
1F4S
 
 | | STRUCTURE OF TRANSCRIPTIONAL FACTOR ALCR IN COMPLEX WITH A TARGET DNA | | Descriptor: | DNA (5'-D(P*CP*GP*TP*GP*CP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*GP*CP*AP*CP*G)-3'), ETHANOL REGULON TRANSCRIPTIONAL FACTOR, ... | | Authors: | Cahuzac, B, Cerdan, R, Felenbok, B, Guittet, E. | | Deposit date: | 2000-06-09 | | Release date: | 2001-09-28 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of an AlcR-DNA complex sheds light onto the unique tight and monomeric DNA binding of a Zn(2)Cys(6) protein.
Structure, 9, 2001
|
|
1F5E
 
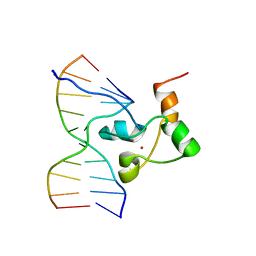 | | STRUCTURE OF TRANSCRIPTIONAL FACTOR ALCR IN COMPLEX WITH A TARGET DNA | | Descriptor: | DNA (5'-D(P*CP*GP*TP*GP*CP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*GP*CP*AP*CP*G)-3'), ETHANOL REGULON TRANSCRIPTIONAL FACTOR, ... | | Authors: | Cahuzac, B, Cerdan, R, Felenbok, B, Guittet, E. | | Deposit date: | 2000-06-14 | | Release date: | 2001-09-28 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | The solution structure of an AlcR-DNA complex sheds light onto the unique tight and monomeric DNA binding of a Zn(2)Cys(6) protein.
Structure, 9, 2001
|
|
1TUJ
 
 | | Solution structure of the honey bee general odorant binding protein ASP2 in complex with trimethylsilyl-d4 propionate | | Descriptor: | 3-TRIMETHYLSILYL-PROPIONATE-2,2,3,3,-D4, odorant binding protein ASP2 | | Authors: | Lescop, E, Briand, L, Pernollet, J.-C, Guittet, E. | | Deposit date: | 2004-06-25 | | Release date: | 2005-09-20 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the honey bee general odorant binding protein ASP2 in complex with trimethylsilyl-d4 propionate
To be Published
|
|
1R1B
 
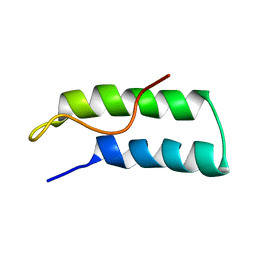 | | EPRS SECOND REPEATED ELEMENT, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRNA SYNTHETASE | | Authors: | Cahuzac, B, Berthonneau, E, Birlirakis, N, Mirande, M, Guittet, E. | | Deposit date: | 1998-12-15 | | Release date: | 1999-12-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A recurrent RNA-binding domain is appended to eukaryotic aminoacyl-tRNA synthetases.
EMBO J., 19, 2000
|
|
1MFK
 
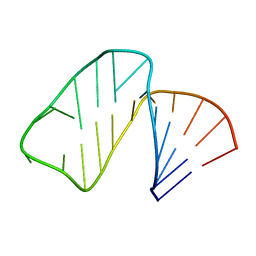 | | Structure of Prokaryotic SECIS mRNA Hairpin | | Descriptor: | 5'-R(P*GP*GP*CP*GP*GP*UP*UP*GP*CP*AP*GP*GP*UP*CP*UP*GP*CP*AP*CP*CP*GP*CP*C)-3' | | Authors: | Fourmy, D, Guittet, E, Yoshizawa, S. | | Deposit date: | 2002-08-12 | | Release date: | 2002-11-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of Prokaryotic SECIS mRNA Hairpin and its Interaction with Elongation Factor SELB
J.Mol.Biol., 324, 2002
|
|
1ZC5
 
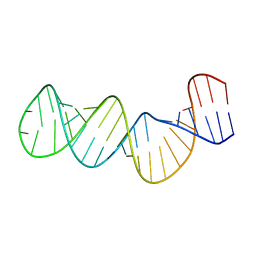 | | Structure of the RNA signal essential for translational frameshifting in HIV-1 | | Descriptor: | HIV-1 frameshift RNA signal | | Authors: | Gaudin, C, Mazauric, M.H, Traikia, M, Guittet, E, Yoshizawa, S, Fourmy, D. | | Deposit date: | 2005-04-11 | | Release date: | 2005-06-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the RNA Signal Essential for Translational Frameshifting in HIV-1
J.Mol.Biol., 349, 2005
|
|
2ALC
 
 | | ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR DNA-BINDING DOMAIN FROM ASPERGILLUS NIDULANS | | Descriptor: | PROTEIN (ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR), ZINC ION | | Authors: | Cerdan, R, Cahuzac, B, Felenbok, B, Guittet, E. | | Deposit date: | 1999-01-20 | | Release date: | 2000-01-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of AlcR (1-60) provides insight in the unusual DNA binding properties of this zinc binuclear cluster protein.
J.Mol.Biol., 295, 2000
|
|
1SQK
 
 | | CRYSTAL STRUCTURE OF CIBOULOT IN COMPLEX WITH SKELETAL ACTIN | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hertzog, M, Van Heijenoort, C, Didry, D, Gaudier, M, Gigant, B, Coutant, J, Didelot, G, Preat, T, Knossow, M, Guittet, E, Carlier, M.F. | | Deposit date: | 2004-03-19 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The beta-Thymosin/WH2 Domain; Structural Basis for the Switch from Inhibition to Promotion of Actin Assembly
Cell(Cambridge,Mass.), 117, 2004
|
|
1MT4
 
 | | Structure of 23S ribosomal RNA hairpin 35 | | Descriptor: | 23S ribosomal Hairpin 35 | | Authors: | Lebars, I, Yoshizawa, S, Stenholm, A.R, Guittet, E, Douthwaite, S, Fourmy, D. | | Deposit date: | 2002-09-20 | | Release date: | 2003-01-14 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of 23S rRNA hairpin 35 and its interaction with the tylosin-resistance methyltransferase RlmAII
Embo J., 22, 2003
|
|
